Abstract
COVID-19 has become a global challenge as there are very few treatment options available. This has proved to impact several physiological implications like immunological injury, myocardial infarction, micro-thrombus formation, neurological complications and multi-organ dysfunction. A combination therapy or a systems pharmacology approach can be adopted to fight against COVID-19. Here, we have proposed withaferin A as a system pharmacophore employing molecular docking strategy using AutoDock Vina and utilising different bioinformatics tools like PharmMapper, STRING database and PANTHER Pathway enrichment analysis. Docking results show that withaferin A exhibits a significant binding affinity with P2Y12 receptor, vitamin D-binding protein and annexin A5, hence implying that it could play a role in anti-thrombosis. Protein–protein interaction network showed its importance in innate immune system. Results also show that this molecule may have significant potential to modulate T cell activation too. Text mining results showed association of STAT3 with withaferin A. Our studies propose that withaferin A might also conquer the cytokine storm via STAT3. This study concludes that two strong targets of withaferin A, i.e. vitamin D-binding protein and STAT3, have been identified and that withaferin A can be used as a system pharmacophore for drug development in order to combat COVID-associated complicacies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
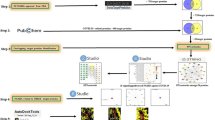
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has severely impacted the global health economics. Also, as this is a recently emerging disease, the available treatment options are less. Though respiratory failure and acute pneumonia are the main causes of the complication, COVID-19 has also proved to imply immunological injury, myocardial infarction, micro-thrombus formation and multi-organ dysfunction [1,2,https://pubchem.ncbi.nlm.nih.gov/) and submitted to PharmMapper server, in order to identify its possible drug targets. The target information of a total of 300 proteins along with their fitness score, pharmacophoric features and the importance of target protein with respect to their relevance in disease conditions was received. They were further arranged in the descending order of their fitness score, and the top 15 targets with a fitness score more than 3.0 were considered for further studies. It was observed that considerable number of targets in the top was of importance to cancer disease. Since our study was focussed on finding the activity of withaferin A in different pathways other than cancer, the top 15 targets were selected (Table 1) based on their relevance in immune system and thrombolytic effects of human body.
Molecular Docking Studies
The crystal structures of the targets were downloaded in PDB format from Protein Data Bank (http://www.rcsb.org/pdb) using their respective PDB-IDs. The peptide chains associated with the co-crystallised compounds were segregated from the respective targets. The co-crystallised ligands were then processed from PDB to their PDBQT formats using AutoDock Tools (ADT) [12].
ADT was used to remove the bound water molecules and lone pairs of electrons, to assign polar hydrogens and to assign Kollman and Gasteiger charges to the protein structures. The prepared protein targets were saved in PDBQT formats by AutoDock. The grid centre coordinates were computed based on the coordinates of existing bound co-crystallised ligand, using the Graphical User Interface of BIOVIA Discovery Studio [13]. The information of grid centre, target protein details and ligand details were collated in a configuration file. AutoDock Vina was used for docking [14], using the information from the configuration file, by using Python script [15]. The co-crystallised compounds and withaferin A were docked at the ligand binding sites of the corresponding targets using AutoDock Vina, kee** the protein as rigid and the ligand as flexible. AutoDock evaluates the protein–ligand interaction affinities by calculating the binding free energy (∆G). The docked structures were visualised in Biovia Discovery Studio. Considering the binding poses of ligand and its interaction with the target protein, the docked structures with the most energetically favourable conformations were considered. The visualisation gave an insight into the ligand binding pocket, interacting amino residues in terms of hydrogen bond interaction and hydrophobic interactions.
Protein–Protein Interaction Network
STRING is an online database (https://string-db.org/) of known and predicted protein–protein interactions. These interactions include direct (physical) and indirect (functional) associations; they stem from computational prediction, from knowledge transfer between organisms and from interactions aggregated from other (primary) databases [16]. The 300 targets retrieved from PharmMapper served as seed protein inputs in STRING database. With the given list of input proteins, STRING can generate the associated PPI networks and pathways affected by withaferin A.
Pathway Enrichment Analysis
PANTHER Pathway (www.pantherdb.org) consists of over 177 pathways, primarily signalling pathways, each with subfamilies and protein sequences mapped to individual pathway components [17]. The targets received as a result of query given to PharmMapper, served as list of input proteins for analyses in PANTHER Pathway. PANTHER classification system was used to understand the most enriched pathways that occurred among the set of protein.
Text Mining
In order to further substantiate our study on role of withaferin A, in relation to combating COVID-19 complications, we employed two R packages, “pubmed.mineR” and “easyPubMed” [18, 19] to carry out text mining. “gene_atomization” automatically fetched the genes (HGNC approved Symbol) from the text and report their frequencies. The gene name and their frequencies are shown in Fig. 5.
Results
Targets of Thrombosis and Withaferin A
There are two types of antithrombotic agents, i.e. antiplatelet agents and anticoagulants. Antiplatelet agents work in three different ways, (i) by preventing the formation of second messengers (thromboxane A2 (TxA2)), (ii) by blocking receptors (P2Y12 and PAR1) involved in platelet activation and (iii) by inhibiting platelet aggregation by binding to integrin αIIbβ3. Anticoagulant agents work by either inhibiting post-translational modification of coagulation proteins, such as factor Xa (FXa) and prothrombin, or inhibiting FXa and thrombin directly [20]. Studies have shown that withaferin A exhibited inhibition in thrombin-catalysed fibrin polymerisation and platelet aggregation, in wet lab experiments. It also showed inhibition in the activities and production of thrombin and FXa [21].
The molecular interactions and targets of withaferin A are still unknown. To elucidate the mechanism of withaferin A and its role in cardiovascular disease, in our previous papers, we screened 6 targets using molecular docking and found HMG-CoA reductase and angiotensin-converting enzyme (ACE2) as suitable targets. In the current study, we have extended our findings to search for antithrombotic effect. P2Y1 and P2Y12 receptors are responsible for platelet aggregation, where P2Y1 initiates and P2Y12 completes the process [22]. Hence, for better insight, we have carried out molecular docking studies of withaferin A with P2Y1 and P2Y12 receptor (Fig. 1), in this article. It is observed that withaferin A bound with P2Y1 has an interaction energy of − 9.5 kcal/mol, whereas the interaction energy of its own crystallised ligand was − 8.6 kcal/mol. This seems to be energetically more favourable than the energy with which it binds with its own co-crystallised ligand (MRS2500) (Fig. 1). Docking studies of withaferin A with P2Y12 showed that even with 3 strong H-bonding interactions, the interaction energy was not favourable, as the crystallised ligand bound better with a binding energy of − 10.1 kcal/mol as opposed to − 6.7 kcal/mol of withaferin A. From this, it can be concluded that for the antithrombotic effect of withaferin A to come into play, P2Y12 is not a suitable target. As an extension to this, the 15 targets chosen from results of PharmMapper were scrutinised for any proteins that played any role in thrombotic effects. To compare the efficiencies of those targets in relation to thrombolytic events, we carried out molecular docking of those targets with withaferin A and compared to the binding interaction energy of their corresponding crystallised ligands. The detail results are shown in the Table 1 and Fig. 2a–g. It was found that 2 out of those 15 targets, vitamin D-binding protein and annexin A5, actively had a role to play in anti-thrombosis which is consistent with our findings (Fig. 2a–g).
The 2D interaction diagram of withaferin with targets by Discovery Studio. The targets are as follows: a P2Y12 co-crystalised molecule, interaction energy = − 10.1 kcal/mol. b P2Y12 withaferin A, interaction energy = − 6.7 kcal/mol. c P2Y1 co-crystalised molecule, MRS; interaction energy = − 8.6 kcal/mol. d P2Y1, withaferin A, interaction energy = − 9.5 kcal/mol
The 2D interaction diagram of withaferin A with 7 targets by Discovery Studio. The targets are as follows: a protein interleukin 1, beta convertase. b Migration inhibitory factor protein S100-A9. c Histamine N-methyltransferase. d Leukocyte (neutrophil) elastase. e Vitamin D-binding protein. f Annexin A5. g Alpha1-Antitrypsin
Molecular Targets of Withaferin A and Protein–Protein Interaction Network
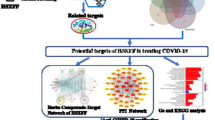
The molecular targets of withaferin A were predicted by PharmMapper. The PPI network of those molecular targets was analysed by STRING database and is shown in Fig. 3. Detail analysis shows that red nodes are involved in cancer pathway, blue nodes are involved in immune system, and green nodes are involved in innate immune system. So, it can be seen that withaferin A acts as an anticancer agent and also protect our immune system.
In this work, we are focused to find out the activity of withaferin A towards different pathways, other than cancer. So, the top 15 targets with a fit score more than 3.0 and those played a role in pathways involving immune system were selected for molecular docking studies, to understand their interaction with withaferin A. Docking results shows that protein interleukin 1-beta convertase, histamine N-methyltransferase, vitamin D-binding protein [23] and annexin A5 are promising targets of withaferin A.
Pathways Affected by Withaferin A
PANTHER Pathway prediction results for withaferin A show 5 major affected pathways. The main pathways are angiogenesis, CCKR signalling, EGFR signalling, gonadotropin-releasing hormone receptor pathway and T cell activation pathway (Fig. 4). Further studies focussing on aspects of T cell activation pathways can lead to solutions in combating COVID-19 associated complicacies.
Text Mining
After text mining, the PubMed literature search with the keyword “withaferin A” showed the top 50 gene names along with their term frequencies (Fig. 5). Analysis showed that the biological process and molecular function of those genes belong to cytokines, involved in immune response and chemokine receptor binding. This is indicative of a possibility of withaferin A may help to combat the dreaded cytokine storm, observed in persons affected by COVID-19.
Discussion
The process of formation of blood clot is termed as thrombosis. This clot can block or obstruct blood flow in the blood vessel. Clinical reports provide evidence that vitamin D deficiency increases the probability of thrombotic episodes. So, vitamin D and its associated molecules play role in the regulation of thrombosis-related pathways. Vitamin D binds to vitamin D-binding protein (VDBP) to get carried to all vital target organs, where it serves as a natural ligand to vitamin D receptors, for enabling their biological actions [23]. COVID-19 patients have demonstrated severe endothelial injury and widespread vascular thrombosis. However, it was reported that VDBP might also play a critical role in severe pulmonary complications of COVID-19. In acute respiratory distress syndrome (ARDS), at sites of endothelial injury, VDBP release promotes monocyte and neutrophil attraction, aggregation and activation to generate an oxidative burst. Thus, it can be seen that VDBP is a valid target candidate in COVID-19. By competing for the same binding site on VDBP, withaferin A may help aid in curbing disease progression. The blocking of either the P2Y1 or the P2Y12 receptor seems to be equally effective in in vivo conditions, as far as both platelet consumption and thrombin generation are concerned [20]. Published literature shows that P2Y1 receptor plays a role in both arterial and venous thrombosis [20, 24, 25]. Withaferin A has shown binding efficacy with P2Y1 receptor but not with P2Y12, but there are still chances that it would exhibit certain level of antithrombotic effects.
The identified targets, protein interleukin 1-beta convertase and histamine n-methyltransferase, are known to exhibit neuromodulatory effects. Tissue damage is an outcome of pathogen invasion, and inflammation is a biological response to it. These proteins are known to play a role as neuromodulator that aids in cellular defence and tissue repair in most tissues. Studies have confirmed that COVID-19 causes certain symptoms classified as neurological signs. The pathological sequence includes nausea, damaged respiratory tract, anosmia, ischemic strokes, cerebral infarction and headaches in a generation of younger patients, without any comorbidity. These symptoms indicate that COVID-19 infection affects the central nervous system [9, 10]. Our study has identified two targets, against whom withaferin A can be employed, to combat the neurological injury inflicted by the virus infection.
PPI network obtained from STRING database depicts that withaferin A has anticancer activity. This has also been established by several experiments. From the PPI network, it is also found that it has high impact on immune system and innate immune system. We encounter millions of pathogens in our day-to-day life, either through direct contact or inhalation. Our innate immune system protects us from the infection, caused by those pathogens, during the initial critical hours to the first few days of exposure to a new pathogen. Given its mode of action, it definitely plays a central role in combating this novel COVID-19 virus [26, 27]. In the current study, the PANTHER Pathway enrichment analysis shows the involvement of T cell activation pathway, along with other pathways operating in cancer disease. Studies have shown that T cells are deregulated in COVID-19, but their exact molecular features are yet to be fully understood [28]. A few researches have shown that hyper activated T cells potentially contribute to T cell deregulation and immune-mediated tissue destruction (immunopathology) in COVID-19. PANTHER Pathway gene enrichment result shows that it has an impact on T cell activation. But it is not clear whether withaferin A will trigger T cells or modulate its activation. Further studies are required to conclude anything about T cell activation or modulation by withaferin A.
We also carried out text mining on withaferin A which further supports our findings. Text mining shows that most data has been found for STAT3 and withaferin A. STAT3 is associated with autoimmune disorder. There might be a possibility that withaferin A could operate through STAT3 also. Though most of the genes retrieved from the result were related to cancer, substantial number of genes was associated with chemokine and cytokines [29]. Chemokine and cytokine activation leads to inflammation that occurs due to exposure to harmful stimuli, such as microbial pathogens [30, 31]. Our study suggests that withaferin A might play a role in either conquering or lessening of the intensity of the cytokine storm, observed in affected people. Thorough studies on withaferin A administration are required to conclude about its role in combating cytokine storm.
Conclusion
Our findings provide data supporting on how withaferin A may have a systemic role in overcoming several immunosuppressive events, occurring as a result of COVID-19. Our studies also suggest that withaferin A has some role to act as an antithrombotic agent.
Our current study suggests that this system pharmacophore can be used to develop a promising drug for COVID-19, kee** in mind the role of withaferin A in immune system and thrombosis.
Data Availability
The data supporting the findings of this study are available in the article.
References
Price, L. C., McCabe, C., Garfield, B., & Wort, S. J. (2020). Thrombosis and COVID-19 pneumonia: The clot thickens! The European Respiratory Journal, 56, 2001608.
Chen, W., & Pan, J. Y. (2021). Anatomical and pathological observation and analysis of SARS and COVID-19: Microthrombosis is the main cause of death. Biological Procedures Online, 23, 4.
Wu, Y., Huang, X., Sun, J., **e, T., Lei, Y., Muhammad, J., Li, X., Zeng, X., Zhou, F., Qin, H., Shao, L., & Zhang, Q. (2020). Clinical characteristics and immune injury mechanisms in 71 patients with COVID-19. mSphere, 5, e00362-20.
Bonow, R. O., Fonarow, G. C., O’Gara, P. T., & Yancy, C. W. (2020). Association of coronavirus disease 2019 (COVID-19) with myocardial injury and mortality. JAMA Cardiology, 5, 751–753.
Ravindran, R., Sharma, N., Roy, S., Thakur, A. R., Ganesh, S., Kumar, S., Devi, J., & Rajkumar, J. (2015). Interaction studies of Withania somnifera’s key metabolite withaferin A with different receptors assosciated with cardiovascular disease. Current Computer-Aided Drug Design, 11, 212–221.
Acharya, P., Huded, P., Bettadahalli, S., Zarei, M., Uppin, V., Venugopal, N., & Talahalli, R. R. (2020). Withaferin-A down-regulate enterohepatic circulation of bile acids: An insight from a hyperlipidemic rat model. Journal of Agriculture and Food Research, 2, 100035.
Trivedi, M. K., Gangwar, M., Mondal, S. C., & Jana, S. (2017). Immunomodulatory properties and biomarkers characterization of novel Withania somnifera based formulation supplemented with minerals in Sprague Dawley rats. Oriental Pharmacy and Experimental Medicine, 17, 59–69.
Turner, M. D., Nedjai, B., Hurst, T., & Pennington, D. J. (2014). Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochimica et Biophysica Acta, 1843, 2563–2582.
Hewett, S. J., Jackman, N. A., & Claycomb, R. J. (2012). Interleukin-1β in central nervous system injury and repair. European Journal of Neurodegenerative Disease, 1, 195–211.
Jarrahi, A., Ahluwalia, M., Khodadadi, H., da Silva Lopes Salles, E., Kolhe, R., Hess, D. C., Vale, F., Kumar, M., Baban, B., Vaibhav, K., & Dhandapani, K. M. (2020). Neurological consequences of COVID-19: What have we learned and where do we go from here? Journal of Neuroinflammation, 17, 286.
Wang, X., Shen, Y., Wang, S., Li, S., Zhang, W., Liu, X., Lai, L., Pei, J., & Li, H. (2017). PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research, 45, W356–W360.
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30, 2785–2791.
BIOVIA, Dassault Systèmes, [Disovery studio], [4.0], San Diego: Dassault Systèmes 2012. Available from: https://discover.3ds.com/discovery-studio-visualizer-download. Accessed 25 July 2021.
Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31, 455–461.
Sanner, M. F. (1999). Python: A programming language for software integration and development. Journal of Molecular Graphics & Modelling, 17, 57–61.
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., Simonovic, M., Doncheva, N. T., Morris, J. H., Bork, P., Jensen, L. J., & Mering, C. V. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47, D607–D613.
Mi, H., & Thomas, P. (2009). PANTHER Pathway: An ontology-based pathway database coupled with data analysis tools. Methods in Molecular Biology (Clifton, N.J.), 563, 123–140.
Rani, J., Shah, A. B., & Ramachandran, S. (2015). pubmed.mineR: An R package with text-mining algorithms to analyse PubMed abstracts. Journal of Biosciences, 40, 671–682.
Damiano, A., Maintainer, F. and Fantini, D. (2019). Package “easyPubMed,” https://cran.r-project.org/web/packages/easyPubMed/easyPubMed.pdf
Rhodes, J. M., Subramanian, S., Laird, E., Griffin, G., & Kenny, R. A. (2021). Perspective: Vitamin D deficiency and COVID-19 severity - Plausibly linked by latitude, ethnicity, impacts on cytokines, ACE2 and thrombosis. Journal of Internal Medicine, 289, 97–115.
Ku, S. K., & Bae, J. S. (2014). Antiplatelet, anticoagulant, and profibrinolytic activities of withaferin A. Vascular Pharmacology, 60, 120–126.
Hechler, B., & Gachet, C. (2011). P2 receptors and platelet function. Purinergic Signalling, 7, 293–303.
Speeckaert, M. M., Speeckaert, R., & Delanghe, J. R. (2020). Vitamin D binding protein in COVID-19. Clinical Medicine (London, England), 20, e136–e137.
Lenain, N., Freund, M., Léon, C., Cazenave, J. P., & Gachet, C. (2003). Inhibition of localized thrombosis in P2Y1-deficient mice and rodents treated with MRS2179, a P2Y1 receptor antagonist. Journal of Thrombosis and Haemostasis: JTH, 1, 1144–1149.
Baurand, A., & Gachet, C. (2003). The P2Y(1) receptor as a target for new antithrombotic drugs: A review of the P2Y(1) antagonist MRS-2179. Cardiovascular Drug Reviews, 21, 67–76.
Schultze, J. L., & Aschenbrenner, A. C. (2021). COVID-19 and the human innate immune system. Cell, 184, 1671–1692.
Chowdhury, M. A., Hossain, N., Kashem, M. A., Shahid, M. A., & Alam, A. (2020). Immune response in COVID-19: A review. Journal of Infection and Public Health, 13, 1619–1629.
Kalfaoglu, B., Almeida-Santos, J., Tye, C. A., Satou, Y., & Ono, M. (2021). T-cell dysregulation in COVID-19. Biochemical and Biophysical Research Communications, 538, 204–210.
Pum, A., Ennemoser, M., Adage, T., & Kungl, A. J. (2021). Cytokines and chemokines in SARS-CoV-2 infections-therapeutic strategies targeting cytokine storm. Biomolecules, 11, 91.
Khalil, B. A., Elemam, N. M., & Maghazachi, A. A. (2021). Chemokines and chemokine receptors during COVID-19 infection. Computational and Structural Biotechnology Journal, 19, 976–988.
Coperchini, F., Chiovato, L., Ricci, G., Croce, L., Magri, F., & Rotondi, M. (2021). The cytokine storm in COVID-19: Further advances in our understanding the role of specific chemokines involved. Cytokine & Growth Factor Reviews, 58, 82–91.
Funding
Dr. Sujata Roy acknowledges the SERB-TARE fellowship File Number: (TAR/2019/000062).
Author information
Authors and Affiliations
Contributions
Sujata Roy: writing, conceptualisation, methodology, software, editing and reviewing.
Ashasmita S. Mishra: molecular docking, protein–protein interaction and pathway analysis, writing and editing.
Bhuvaneswari Varadarajan: text mining.
Srayaa Sathish: pathway analysis, writing and editing.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors declare their consent to publish their work.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mishra, A.S., Varadarajan, B., Sathish, S. et al. Withaferin A for COVID-19: a Network Pharmacology Approach. Appl Biochem Biotechnol 195, 4983–4994 (2023). https://doi.org/10.1007/s12010-023-04525-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04525-7