Abstract
There are only a few models developed for risk-stratifying COVID-19 patients with suspected pneumonia in the emergency department (ED). We aimed to develop and validate a model, the COVID-19 ED pneumonia mortality index (CoV-ED-PMI), for predicting mortality in this population. We retrospectively included adult COVID-19 patients who visited EDs of five study hospitals in Texas and who were diagnosed with suspected pneumonia between March and November 2020. The primary outcome was 1-month mortality after the index ED visit. In the derivation cohort, multivariable logistic regression was used to develop the CoV-ED-PMI model. In the chronologically split validation cohort, the discriminative performance of the CoV-ED-PMI was assessed by the area under the receiver operating characteristic curve (AUC) and compared with other existing models. A total of 1678 adult ED records were included for analysis. Of them, 180 patients sustained 1-month mortality. There were 1174 and 504 patients in the derivation and validation cohorts, respectively. Age, body mass index, chronic kidney disease, congestive heart failure, hepatitis, history of transplant, neutrophil-to-lymphocyte ratio, lactate dehydrogenase, and national early warning score were included in the CoV-ED-PMI. The model was validated with good discriminative performance (AUC: 0.83, 95% confidence interval [CI]: 0.79–0.87), which was significantly better than the CURB-65 (AUC: 0.74, 95% CI: 0.69–0.79, p-value: < 0.001). The CoV-ED-PMI had a good predictive performance for 1-month mortality in COVID-19 patients with suspected pneumonia presenting at ED. This free tool is accessible online, and could be useful for clinical decision-making in the ED.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Background
The coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to a global pandemic since December 2019 [1]. The exponential growth of infected individuals, along with 20–30% of in-hospital mortality [2,3,4], has wreaked havoc on global health care systems [2,3,4]. Illness severity for COVID-19 varies drastically between individuals, ranging from asymptomatic or mild illness to severe viral pneumonia with acute respiratory distress or failure [5]. Most patients are categorized into mild-to-moderate COVID-19, defined as subjects with mild symptoms up to mild pneumonia [6]. Nevertheless, decompensation could also afflict these otherwise well-appearing patients rather rapidly due to the difficulty of predicting severe lung injury early in the course of illness [7].
Since the beginning of the COVID-19 pandemic, various prognostic models have been proposed to predict adverse outcomes [8]. However, most of these models were developed for the inpatient setting rather than for the emergency department (ED) [8]. The National Early Warning Score (NEWS) is one of the most accurate tools for detecting patient deterioration outside intensive critical units (ICUs), which is based only on patients’ vital signs [9]. In addition to vital signs, other factors such as age, comorbidities, imaging features and blood tests have also been reportedly associated with COVID-19 severity [8].
Objective
In this study, we attempted to develop and validate a NEWS-based logistic regression model, the COVID-19 ED pneumonia mortality index (CoV-ED-PMI), for adult COVID-19 patients presenting at ED with suspected pneumonia to predict 1-month mortality. Furthermore, we compared CoV-ED-PMI with other recommended prognostic tools [8], including CURB-65, a commonly used risk-stratifying tool for community-acquired pneumonia [10] and 4C Mortality Score, a newly developed tool to predict inpatient mortality related to COVID-19 by the Coronavirus Clinical Characterisation Consortium [11].
Materials and methods
Study setting and design
This retrospective study was conducted in the five study hospitals affiliated with Baylor Scott & White Health (BSWHealth), Texas, USA. This study was performed in accordance with the Declaration of Helsinki amendments. The Institutional Review Board approved this study (reference number: 344143) and waived the requirement for informed consent. The results are reported according to the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) statement [12].
Study population
All consecutive patients visiting the EDs of the study hospitals between 1st March and 30th November 2020 were screened. Patients fulfilling the following criteria were eligible for inclusion: (1) tested positive for SARS-CoV-2 by quantitative reverse transcriptase polymerase chain reaction (RT-PCR) from samples collected through nasopharyngeal or oropharyngeal swabs; (2) age ≥ 18 years; (3) received chest X-ray (CXR) examination for suspected pneumonia. All patients were followed up for 1 month. If a single patient visited EDs in these study hospitals multiple times, only the data of the first visit were extracted for analysis. Because of the limited capacity for quantitative RT-PCR testing, during March and April 2020, SARS-CoV-2 screening was restricted to patients with contact or travel history or patients with suspicious laboratory or imaging findings. Since May 2020, the decision to have the RT-PCR test was left at the discretion of the ED clinicians without further limitation. The CXR examination was ordered under clinical suspicion for pneumonia. During the study period, the vaccination for COVID-19 was not yet available.
Data collection and outcome measures
All potential candidate variables were extracted from the BSWHealth’s electronic medical record (EMR) system (Epic, Verona, WI). We used previous reports [13] to assist us in selecting promising candidate predictors, including basic demographics, comorbidities documented through diagnostic codes linked to ambulatory primary care and specialty encounters, body mass index (BMI) (kg/m2) recorded within the previous 12 months before the index ED visit, presenting vital signs and supplemental oxygen use recorded at ED triage, the first CXR report during the index ED visits, and the blood test results. Since our study design was retrospective in nature, the descriptive terms used by radiologists were not standardized. To account for CXR findings in the analysis, we first randomly selected 10% of the available CXR reports. Second, we recorded the descriptive or diagnostic terms used by radiologists in the pilot-run extraction form and calculated the appearance frequency of these terms. Finally, we listed those terms with appearance frequency above 5% in the formal extraction form for research assistants blinded to the patient outcomes to extract. Neutrophil-to-lymphocyte ratio (NLR) was calculated by dividing the percentage of neutrophils by percentage of lymphocytes. If the percentage of lymphocytes was zero, the value would be replaced by 0.5. The NEWS [14], CURB-65 [10] and 4C Mortality Score [11] were computed according the variables recorded at ED triage.
We specified 1-month mortality as the primary outcome, defined as all-cause mortality occurring within 1 month after the index ED visit. We checked the survival status of all included patients through the interconnected EMR system in BSWHealth. Therefore, even if the death occurred at non-study hospitals, the death record could still be identified in the EMR system.
Sample size
Because of the retrospective nature of this study, the number of eligible patients during the study period determines the final sample size. Samples were split into the derivation and validation cohorts at a 70-to-30 ratio chronologically to simulate a prospective validation study.
Statistical analysis
Categorical variables are presented as counts with proportions, and continuous variables are presented as medians with interquartile range. Differences between groups were evaluated by chi-square test for categorical variables or Wilcoxon’s rank-sum test for continuous variables.
In the derivation cohort, we performed the multivariable logistic regression analyses to estimate the associations between candidate variables and outcome. We placed all candidate variables in the regression model for variable selection regardless of their significance by univariate analyses. We employed generalized additive models (GAMs) [15] to explore non-linear effects of the continuous variables on outcomes and to identify the optimal cut-off points to transform these variables into categorical variables. We developed the final prediction model by stepwise variable selection procedure with iterations. We defined the significance levels for entry and to stay at 0.15 to avoid exclusion of potential variables. The final prediction model was derived by excluding non-significant variables sequentially until all regression coefficients were significant.
Missing values in variables like BMI were considered as missingness at random and replaced with imputed values based on age and sex by simple linear regression method. For missing values of missingness not at random like blood tests, we set a binary variable for each blood test to indicate the presence or absence of missing values. Then, this indicator variable was multiplied by the variable of blood test and used during the model-building process.
In the validation cohort, we assessed the discriminative performance of the derived model by area under the receiver operating characteristic (ROC) curve (AUC). We evaluated model calibration by the Hosmer–Lemeshow goodness-of-fit test and a calibration plot to compare the prediction with the observed risk of 1-month mortality. We also validated the CURB-65 and 4C Mortality Score by calculating the AUC in the validation cohort. The discriminative performance of the CoV-ED-PMI, CURB-65, and 4C Mortality Score were compared by DeLong test [16].
We used R 4.0.3 software (R Foundation for Statistical Computing, Vienna, Austria) to analyze data by using packages of My.stepwise, VGAM, ROCR, pROC, gbm and ResourceSelection. A two-tailed p-value < 0.05 was considered statistically significant.
Results
The patient selection process is shown in Fig. 1. The final cohort included a total of 1678 patients. Of them, the derivation cohort consisted of 1,174 patients dated before 15th July, and the validation cohort 504 patients dated after 16th July 2020.
The characteristics of the patients and all candidate variables in the final cohort, and their comparisons between the derivation and validation cohorts are presented in Table 1. Overall, the median patient age was 54.9 years, and 840 patients (50.1%) were male. Pneumonia was diagnosed in 925 patients (55.1%) by CXR. The median NEWS, CURB-65 and 4C Mortality Score were 3, 0, and 5, respectively. The median number of missing values per patient was 1, and 867 patients (51.6%) had at least one missing value. A total of 180 patients (10.7%) died 1 month after the index ED visit. Several differences existed between the derivation and validation cohorts, resulting in higher mortality in the validation cohort.
The differences between patients stratified by 1-month mortality in the derivation cohort are shown in Table 2. For the derivation cohort, the GAM plots illustrated the association of logit (p), where p represented the probability for 1-month mortality, with age, BMI, NLR, lactate dehydrogenase (LDH), and NEWS (Supplemental Fig. 1). If logit (p) was greater than zero, the odds for sustaining 1-month mortality would be greater than one. Therefore, 27.5 was selected as a cut-off point to transform BMI into a categorical variable used in logistic regression analysis.
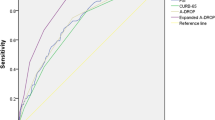
As shown in Table 3, the CoV-ED-PMI included nine variables, resulting in excellent discriminatory performance in the derivation cohort (AUC: 0.94, 95% confidence interval [CI]: 0.92–0.96). The CoV-ED-PMI has been launched online as the following URL: https://chou2.chou-tw.com/index.php/predict/). In the validation cohort, the CoV-ED-PMI also demonstrates good discriminatory performance (AUC: 0.83, 95% CI: 0.79–0.87). Nonetheless, the calibration plot (Fig. 2) indicates that the CoV-ED-PMI may overestimate and underestimate mortality for patients higher and lower than predicted mortality of approximately 40%, respectively (Hosmer–Lemeshow test p-value: < 0.001). In the validation cohort, the discriminatory performance of the CoV-ED-PMI was significantly better than CURB-65 (AUC: 0.74, 95% CI: 0.69–0.79, p-value: < 0.001); in contrast, the CoV-ED-PMI did not significantly outperform 4C Mortality Score (AUC: 0.81, 95% CI: 0.77–0.86, p-value: 0.30) (Fig. 3).
Discussion
Main findings
We developed and validated a logistic regression model, the CoV-ED-PMI, to assist physicians in estimating the probability of 1-month mortality of COVID-19 patients assessed in the ED with suspected pneumonia, aiming to assist the clinician in the safe disposition of these patients. This model included nine common variables and achieved good discriminative performance in the validation cohort. This model outperformed CURB-65 and was similar to 4C Mortality Score in discriminative performance.
Comparison with previous studies
After reviewing more than 100 prognostic models, Wynants et al. [8] indicated that CURB-65 and 4C Mortality Score were 2 of the most recommended tools in predicting mortality for inpatients with COVID-19. The highest AUC achieved by CURB-65 in previous external validation studies for inpatients with COVID-19 was 0.84 [17]. However, in our validation cohort, the discriminatory performance of CURB-65 was just 0.74. The different case mix between hospitalized patients and patients presenting at ED may render CURB-65 unfit for use in the ED. As for 4C Mortality Score [11], which was specifically designed for COVID-19, its discriminative performance in our validation cohort (AUC: 0.81) was even higher than its performance in the original study (AUC: 0.77). As a result, although 4C Mortality Score study [11] enrolled patients hospitalized for high likelihood of contracting SARS-CoV-2 between February and June 2020 without confirmation by RT-PCR, being quite different from our cohort, the results of current study suggested that 4C Mortality Score may also be applied to COVID-19 patients presenting to ED with suspected pneumonia.
Interpretation of current analysis
Distinct from most predictive models that analyze each vital sign separately, the CoV-ED-PMI employs a composite score of vital signs, i.e., NEWS, to assist in risk stratification for COVID-19 patients presenting at the ED with suspected pneumonia. For general ED patients, NEWS had been shown to accurately predict both in-hospital mortality and ICU admission [9]. For COVID-19 patients, Covino et al. [18] indicated that NEWS was the most accurate early warning score in predicting ICU admission within 48 h and 7 days from ED admission. We selected NEWS as the foundation of CoV-ED-PMI not only because of emergency physician (EP) familiarity with the scoring tool, but also for its validated prognostic performance. However, given that NEWS only uses initial vital signs assessed at ED triage, it is less suited to predict the full trajectory over a period of 1 month. As such, we added variables commonly available at ED visits to improve the prognostic performance of NEWS.
Many studies have demonstrated that age and comorbidities are associated with mortality of COVID-19 [13]. Similarly, in CoV-ED-PMI, age, congestive heart failure, chronic kidney disease, hepatitis, and history of transplantation were identified as significant predictors. Besides these comorbidities, several studies have indicated the association between obesity and increased risk of mechanical ventilation, severe pneumonia, and death with COVID-19 [19, 20]. Obesity was also used as a predictor of poor outcome in the 4C mortality score [11]; nonetheless, the thresholds of being obese were not defined explicitly. In our model, the GAM plot revealed that not only obesity (BMI > 30 kg/m2), but also the upper range of overweight (BMI between 27.5 and 30 kg/m2) may be associated with increased 1-month mortality.
Activation of multiple inflammatory pathways has been noted in COVID-19 patients [21]. In CoV-ED-PMI, NLR and LDH were selected as the representative biomarkers of inflammation. Patients with severe COVID-19 may present with lymphopenia [22]. Therefore, NLR could indicate both the general inflammatory status and the underlying severity of COVID-19. Normal ranges of the NLR has been reported to be between 0.78 and 3.53 [23] but no studies reported an absolute value of NLR to define severity of COVID-19. As shown in Supplemental Fig. 1, after logit transformation, there was a near-linear association between NLR and the probability of 1-month mortality. Therefore, NLR was treated as a continuous variable in our model. On the other hand, LDH is a ubiquitous enzyme found in nearly all living cells, which is released during tissue injury, and thus its plasma concentration increases during various pathologic processes. For COVID-19, LDH has also been reported as a prognostic biomarker [24]. In our analysis, after logit transformation, LDH concentrations are also proportional to the probability of 1-month mortality (Supplemental Fig. 1) and thus also treated as continuous variable in our model.
Interestingly, no CXR features were identified to be a significant predictor in CoV-ED-PMI. CXR severity score, quantified by counting the involved lung areas, was reported to be predictive of risk for hospital admission and intubation in COVID-19 patients [25]. In contrast, in our study, only the qualitative data, i.e., findings in the radiologists’ reports, were analyzed and the CXR report forms were not prospectively designed with uniform definitions, which may introduce heterogeneity, leading to the null association between CXR features and outcomes.
Future implications
The COVID-19 pandemic is threatening the capacity of EDs around the world. A better understanding of the factors associated with the disease severity can help withhold the spread of the pandemic. Approximately 81% of COVID-19 patients were categorized as mild-to-moderate severity (mild symptoms up to mild pneumonia) [6], and most of them did not need specific treatments and hospitalization [26]. Because of the limited hospital capacity, EPs need to stratify patients into different risks categories and identify those with highest risk of disease progression for hospitalization. Currently, EPs are faced with a lack of validated risk assessment tools to assist in the disposition of COVID-19 patients with suspected pneumonia since most previous prediction models [8] were derived from the patient data obtained during the early pandemic. Nevertheless, studies [27] indicate that there was a significant temporal variation in in-hospital mortality of COVID-19, suggesting that early experience in dealing with COVID-19 patients may not be applicable to those diagnosed later. Therefore, we split our cohort chronologically into an earlier derivation cohort and a later validation cohort. The unadjusted mortality of the validation cohort was about twice that of the derivation cohort. This significant difference in mortality may lead to the modest calibration of CoV-ED-PMI in the validation cohort, despite that its good validated discriminative performance. As such, when using CoV-ED-PMI to compute the risk of 1-month mortality, the clinicians should be aware of the potential over- and underestimation.
Most of the previous models [8] enrolled COVID-19 inpatients and aimed to predict the adverse outcomes of these patients post-ED disposition. Conversely, the CoV-ED-PMI we developed is to assist the EPs in pre-disposition assessment and risk stratification. Furthermore, we only focus on those COVID-19 patients with suspected pneumonia because most COVID-19 patients without symptoms or only mild illness can receive minimal, symptomatic treatment [26]. The CoV-ED-PMI only required basic demographics, comorbidities recorded in EHR, vital signs measured at triage and two commonly performed blood tests, which may be computed quite easily by an online calculator. The similar discriminative performance between the CoV-ED-PMI and 4C Mortality Score (which was developed from a large database) may assure EPs of the utility of CoV-ED-PMI in clinical use. However, the effects of vaccination against COVID-19 were not considered in our model. To accommodate these changes, the CoV-ED-PMI needs periodical validation and update to keep up with the rapidly changing pandemic.
Study limitations
First, missing data are common during pandemics of emerging infectious disease because of the uncertainty in standard management, and our study is no exception. Complete case analysis may lead to exclusion of a substantial proportion of the available subjects, thereby leading to a loss of precision and power [28]. To deal with missing data, multiple imputation was applied to replace the missing values given that the missingness was caused by missing completely at random or missing at random [12]. Nonetheless, most missing blood test results in our study were likely due to a lack of clinical indication, which may be associated with the severity of COVID-19 and clinical outcomes. For missing not at random, we used an indicator variable for each variable of blood test. With a better understanding of COVID-19, the indication for blood tests may also change. Nonetheless, we have successfully validated our model with a chronologically split validation cohort, which may mitigate the concerns for the generalizability of our model to some extent. Second, there was no strict definition for COVID-19 patients with suspected pneumonia. Only 55.1% of patients had radiological diagnosis of pneumonia with the remaining diagnosed clinically (Table 1). The different reporting styles of radiologists or the qualitative nature in extraction may in part explain this low proportion. On the other hand, although radiological exams are not the necessary diagnostic criteria for pneumonia [29], when faced with an emerging disease like COVID-19, the agreement in clinical diagnosis of COVID-19-related pneumonia may be susceptible to different diagnostic criteria used by different EPs, which may introduce heterogeneity and influence the applicability of our model. This could only be resolved through a prospective study design with pre-specified diagnostic and inclusion criteria.
Conclusions
The CoV-ED-PMI was developed and validated using clinical data from COVID-19 patients in the ED to predict 1-month mortality. This free-to-use tool has been launched online and can be easily applied in the clinical settings. By entering only ED vital signs and some common variables, it has the potential to help the EPs facilitate the disposition of ongoing COVID-19 patients with suspected pneumonia. Nonetheless, the predicted 1-month mortality is not the only important piece of information in the decision-making process, the final disposition should still be made considering other factors, like the patients’ wish and facilities’ capabilities.
Abbreviations
- COVID-19:
-
Coronavirus disease 2019
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- ED:
-
Emergency department
- NEWS:
-
National Early Warning Score
- ICU:
-
Intensive critical units
- CoV-ED-PMI:
-
COVID-19 ED pneumonia mortality index
- BSWHealth:
-
Baylor scott and white health
- TRIPOD:
-
Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis
- RT-PCR:
-
Reverse transcriptase polymerase chain reaction
- CXR:
-
Chest X-ray
- EMR:
-
Electronic medical record
- BMI:
-
Body mass index
- NLR:
-
Neutrophil-to-lymphocyte ratio
- GAM:
-
Generalized additive model
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the ROC curve
- LDH:
-
Lactate dehydrogenase
- EP:
-
Emergency physician
References
World Health Organization. In: Coronavirus disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed 20 May 2021.
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y et al (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z et al (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062
Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW et al (2020) Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA 323:2052–2059
Wu Z, McGoogan JM (2020) Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 323:1239–1242
Centers for Disease Control and Prevention. In: Interim Clinical Guidance for Management of Patients with Confirmed Coronavirus Disease (COVID-19). 2021. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html. Accessed 20 May 2021.
Bouadma L, Lescure FX, Lucet JC, Yazdanpanah Y, Timsit JF (2020) Severe SARS-CoV-2 infections: practical considerations and management strategy for intensivists. Intensive Care Med 46:579–582
Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328.
Nannan Panday RS, Minderhoud TC, Alam N, Nanayakkara PWB (2017) Prognostic value of early warning scores in the emergency department (ED) and acute medical unit (AMU): a narrative review. Eur J Intern Med 45:20–31
Lim WS, van der Eerden MM, Laing R, Boersma WG, Karalus N, Town GI et al (2003) Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax 58:377–382
Knight SR, Ho A, Pius R, Buchan I, Carson G, Drake TM, et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339.
Collins GS, Reitsma JB, Altman DG, Moons KG (2015) Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 162:55–63
Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J et al (2020) Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect 81:e16–e25
Smith GB, Prytherch DR, Meredith P, Schmidt PE, Featherstone PI (2013) The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation 84:465–470
Hastie TJ, Tibshirani RJ (1990) Generalized additive models. Chapman & Hall/CRC, London
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845
Luo M, Liu J, Jiang W, Yue S, Liu H, Wei S. IL-6 and CD8+ T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight. 2020;5:e139024.
Covino M, Sandroni C, Santoro M, Sabia L, Simeoni B, Bocci MG et al (2020) Predicting intensive care unit admission and death for COVID-19 patients in the emergency department using early warning scores. Resuscitation 156:84–91
Cai Q, Chen F, Wang T, Luo F, Liu X, Wu Q et al (2020) Obesity and COVID-19 severity in a designated hospital in Shenzhen. Chin Diabetes Care 43:1392–1398
Lighter J, Phillips M, Hochman S, Sterling S, Johnson D, Francois F et al (2020) Obesity in patients younger than 60 years is a risk factor for COVID-19 Hospital Admission. Clin Infect Dis 71:896–897
Zeng F, Huang Y, Guo Y, Yin M, Chen X, **ao L et al (2020) Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis 96:467–474
Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y et al (2020) Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan. China Clin Infect Dis 71:762–768
Forget P, Khalifa C, Defour JP, Latinne D, Van Pel MC, De Kock M (2017) What is the normal value of the neutrophil-to-lymphocyte ratio? BMC Res Notes 10:12
Deng X, Liu B, Li J, Zhang J, Zhao Y, Xu K (2020) Blood biochemical characteristics of patients with coronavirus disease 2019 (COVID-19): a systemic review and meta-analysis. Clin Chem Lab Med 58:1172–1181
Toussie D, Voutsinas N, Finkelstein M, Cedillo MA, Manna S, Maron SZ et al (2020) Clinical and chest radiography features determine patient outcomes in young and middle-aged adults with COVID-19. Radiology 297:E197–E206
Centers for Disease Control and Prevention. Therapeutic Management of Adults With COVID-19. 2021. https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/. Accessed 20 May 2021.
Navaratnam AV, Gray WK, Day J, Wendon J, Briggs TWR (2021) Patient factors and temporal trends associated with COVID-19 in-hospital mortality in England: an observational study using administrative data. Lancet Respir Med 9:397–406
Luo X, Zhou W, Yan X, Guo T, Wang B, **a H et al (2020) Prognostic value of C-reactive protein in patients with coronavirus 2019. Clin Infect Dis 71:2174–2179
Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200:e45-e67.
Acknowledgements
The authors acknowledge statistical assistance provided by the Center of Statistical Consultation and Research in the Department of Medical Research, National Taiwan University Hospital.
Funding
This work was supported with grants from the Ministry of Science and Technology, Taiwan for Chih-Hung Wang (109-2634-F-002-041) and Tsung-Chien Lu (109-2823-8-002 -013 –CV). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
EHC, CHW, and TCL contributed to the conception and design of the work. EHC acquired and structured the dataset and performed the analysis. JW, JG, TB and AS contributed to analysis and interpretation of data. CHW, CLT, GOO, and TCL validated and interpreted the results. FYC and CHW contributed to the creation of the online website used in the work. EHC and CHW drafted the manuscript. DH, RR, JD and MHM critically revised the manuscript. TCL supervised the conduction of the wok. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethics approval and consent to participate
This study was approved by the institutional review board of the Baylor Scott & White Research Institute (reference number: 344143) and waived the requirement for informed consent.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the first author (email: eric.chou@bswhealth.org) on reasonable request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chou, E.H., Wang, CH., Chou, FY. et al. Development and validation of a prediction model for estimating one-month mortality of adult COVID-19 patients presenting at emergency department with suspected pneumonia: a multicenter analysis. Intern Emerg Med 17, 805–814 (2022). https://doi.org/10.1007/s11739-021-02882-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11739-021-02882-x