Abstract
The combined use of LiDAR (Light Detection And Ranging) scanning and field inventories can provide spatially continuous wall-to-wall information on forest characteristics. This information can be used in many ways in forest map**, scenario analyses, and forest management planning. This study aimed to find the optimal way to obtain continuous forest data for Catalonia when using kNN imputation (kNN stands for “k nearest neighbors”). In this method, data are imputed to a certain location from k field-measured sample plots, which are the most similar to the location in terms of LiDAR metrics and topographic variables. Weighted multidimensional Euclidean distance was used as the similarity measure. The study tested two different methods to optimize the distance measure. The first method optimized, in the first step, the set of LiDAR and topographic variables used in the measure, as well as the transformations of these variables. The weights of the selected variables were optimized in the second step. The other method optimized the variable set as well as their transformations and weights in one single step. The two-step method that first finds the variables and their transformations and subsequently optimizes their weights resulted in the best imputation results. In the study area, the use of three to five nearest neighbors was recommended. Altitude and latitude turned out to be the most important variables when assessing the similarity of two locations of Catalan forests in the context of kNN data imputation. The optimal distance measure always included both LiDAR metrics and topographic variables. The study showed that the optimal similarity measure may be different for different regions. Therefore, it was suggested that kNN data imputation should always be started with the optimization of the measure that is used to select the k nearest neighbors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Airborne Laser Scanning (ALS) provides spatially continuous data on forest canopies. This data can be used to estimate traditional forest parameters since various metrics calculated from the laser scanning data correlate with the characteristics measured in traditional forest inventories (White et al. 2013). Thanks to these correlations, it is possible to use the ALS data to produce wall-to-wall estimates of those characteristics of forest stands that are often used as input data in forest planning. For example, the lowest heights of the LiDAR (Light Detection And Ranging) echoes mainly represent the elevation of the ground while the highest echoes represent the elevation of the canopy surface. The difference between the highest points classified as vegetation and the ground surface can therefore be used to estimate canopy height. The intermediate echoes provide information on the 3D structural characteristics of trees (Lim et al. 2003; Hyyppä et al. 2008).
Wall-to-wall data are also available on topography. It can be assumed that altitude, slope, and aspect also correlate with the characteristics of forest stands. Especially in forests that have evolved without strong human impact, several forest characteristics systematically change with topography. For example, in the lower lands and pre-Pyrenees of Catalonia (NE Spain), the species composition changes from forests dominated by Pinus halepensis first to P. nigra, then P. sylvestris and eventually to P. uncinata when altitude increases (Rouget et al. 2001).
If there are field-measured data from many sample plots, and the plots represent the whole range of variation in tree stand characteristics, LiDAR and topographic data can be utilized to impute forest data from the field plots to any location of the terrain. The method most used for this purpose is the k-nearest neighbors (kNN) imputation where the data to a certain location are imputed from those k plots that are most similar (“nearest”) to the location in terms of LiDAR metrics (Maltamo et al. 2006) and topographic variables. All variables that are available for both locations, that is, for the field plot as well as for the location to which data are imputed, can be used to assess the similarity between them.
It is possible to impute single variables such as volume, biomass, carbon stock, or sets of variables (Packalen et al. 2012). Imputation of detailed forest inventory data such as the diameter and height distributions per species would make it possible to use the imputed data for all those purposes for which the original forest inventory data are used (Pukkala 2019; Díaz-Yáñez et al. 2020). Examples of these purposes include volume, carbon or biomass estimates, forest planning calculations, and scenario simulations (Trasobares et al. 2022). In addition, wall-to-wall data allow straightforward production of forest maps and visualization of forest data (Jia et al. 2020; Pukkala 2020).
Develo** a measure for assessing the similarity of two locations in a forested landscape consists of the following three sub-tasks: (1) selecting the variables that are used in the similarity measure; (2) selecting the most appropriate transformation for each of these variables; and (3) finding the optimal weights for these variables. In addition, the method for selecting variables for the distance measure and the type of the distance measure needs to be decided (e.g., LeMay and Temesgen 2005; Chirici et al. 2008; Hudak et al. 2008; Latifi et al. 2010).
Packalen et al. (2012) compared three methods for selecting the variables to the distance measure: loadings in canonical correlation (Gittins 1985), random forest importance (Breiman 2001), and combinatorial optimization (i.e., simulated annealing). Of these methods, simulated annealing was the best with a clear margin to the other methods. In the same study, within each method, three distance measures were tested: most similar neighbors (Moeur and Stage 1995; Maltamo et al. 2006), random forest proximity matrix (Crookston and Finley 2008), and optimized weights in Euclidean distance. When five variables were imputed, there were practically no differences in the performance of these three distance measures. The performance criterion in the comparisons of alternative methods was the average of the relative root mean square errors (RMSE) of the imputed variables. When only one variable (dominant height) was imputed, the random forest proximity measure was better than the others. Hudak et al. (2008) found that random forest performed better than most similar neighbors and Euclidean distance when two variables (basal area and tree density) were imputed. The performance of alternative distance measures may depend on the selection of variables that are included in the distance measure (Latifi et al. 2010).
In the present study, the aim was to develop a methodology for imputing forest inventory data from the field plots of the Spanish National Forest Inventory (NFI) to different locations of the Catalan forests (a NE Spanish region). The objective was to develop an imputation method that finds, for a certain location, those NFI plots that have the most similar distribution of tree sizes, growing stock density, and species composition as the location to which data are imputed. Similarity evaluations were based on various percentiles of the diameter distribution of trees, as well as variables that measure stand density and species composition. A similarity measure that minimized the mean of the relative RMSEs of these variables was regarded to be the best (Packalen et al. 2012). In addition, the effect of the number of nearest neighbors (k) that was used in the kNN imputation was analyzed.
Three categories of variables were tested in the similarity assessment: LiDAR metrics, terrain metrics (elevation, slope, aspect), and geographical location (x and y coordinates). Based on Packalen et al. (2012), simulated annealing was used to find the optimal combination of variables for the distance measure. The type of distance measure was weighted Euclidean distance calculated from the selected variables. It was used to find the most similar NFI plots for different locations of a forested landscape.
Materials and methods
Data sources
Two case study areas within Catalonia were used to develop the data imputation method: Solsonès and Alt Urgell counties (41°58′47″ N 1°30′47″ E and 42°14′19″ N 1°24′22″ E, respectively). Inventory plots of the Fourth National Forest Inventory (NFI4) were used as field data. Plots located within the two counties and in a 20-km-wide buffer zone around the counties were used as the imputation dataset. This resulted in 393 plots for Solsonès and 641 plots for Alt Urgell.
Continentality, topography, and latitude combined determine different climatic regions in Catalonia, ranging from Alpine to Mediterranean, but Continental too. Climatic types in Catalonia are driven by average precipitation, seasonal rainfall regime, mean annual temperature, and thermal amplitude, and dictate the overall conditions for tree species to grow. Alt Urgell County is dominated by Pyrenean and Pre-Pyrenean climatic types with rainfall concentrated during the summer season and mean annual precipitation above 1000 and 650 mm, respectively. By contrast, Solsonès County, even if geographically close to Alt Urgell is dominated by dry continental conditions, with mean annual precipitation below 550 mm and thermal amplitude from 17 °C to 20 °C. These conditions favour alpine tree species in Alt Urgell forests, such as Pinus sylvestris (42%) and several other dominant species, while Pinus nigra (50%) dominates Solsonès, followed by Pinus sylvestris (23%) and a clear presence of Quercus species (23%) across the county.
In Catalonia, the fieldwork of NFI4 was carried out between December 2013 and July 2016, and it comprised 5431 plots, of which 93% were also sampled in the previous campaign. In the NFI plots, trees were sampled concentrically. Regeneration (i.e., trees with diameter at breast height (DBH) < 2.5 cm) was assessed in a 5-m radius sub-plot and categorized into four density categories. Trees with 2.5 ≤ DBH < 7.5 cm were also assessed in the 5-m radius sub-plot but only the species, the mean height, and the number of individuals were recorded. In the 5-m, 10-m, 15-m, and 25-m radius sub-plot all trees of 7.5 ≤ DBH < 12.5 cm, 12.5 ≤ DBH < 22.5 cm, 22.5 ≤ DBH < 42.5 cm, and DBH ≥ 42.5 cm, respectively, were measured for DBH and height, the species identified, and quality and stem form categorized (Alberdi et al. 2016). Only individually measured trees (DBH ≥ 7.5 cm) were used in the analyses of this study.
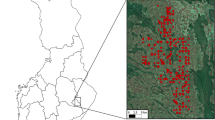
At present, the Cartographic and Geological Institute of Catalonia (ICGC) distributes two LiDAR programs that cover the whole territory of Catalonia. Both coverages were captured with an ALS50-II LiDAR sensor from Leica Geosystems. The second LiDAR coverage, called the LIDARCAT2 project, was flown between 2016 and 2017 with a mean first-return point density of 0.5 pulses·m2. LiDAR data in Solsonès was captured from June to December in 2016 and from May to July in 2017 (Fig. 1). Alt Urgell data were captured from June to December in 2016 and the same period in 2017.
LiDAR data were distributed in blocks of 2 km × 2 km, and their point coordinates were adjusted using longitudinal and transversal flight lines and ground control points. Subsequently, after filtering noisy points, LiDAR returns were automatically classified with Terrascan classification routines (Terrasolid version 017 2017). The automatic classification was started by classifying ground returns. An initial terrain model was built based on the ground classification of the first LiDAR coverage, called LIDARCAT1.
The LIDARCAT1 coverage includes a manual edition of the ground class, the purpose of which was to refine the automatic classification. The ground classification routine molded the model upwards by iteratively adding new laser points to it. Once ground returns were determined, the classification continued with the next classes: low, medium, and high vegetation, buildings, model key points, transmission line poles, and other pillars. After the manual edition, the point cloud was height-normalized by calculating the vertical distance of each return to a Triangular Irregular Model (TIN) generated from returns classified in the model key points class (Blázquez-Casado et al. 2015; Martín-Alcón et al. 2015).
Canopy metrics, such as height statistics to describe the canopy distribution and cover, were calculated from the classified and height-normalized LiDAR point cloud in the location of the NFI4 plots using the USDA Forest Service FUSION v3.2 software (FUSION 2012). The extraction of metrics per plot was based on the maximum radius within which trees were measured in the IFN4 plot. For mature stands (39%), the extraction was done within a 25-m radius, for young stands (49%) in a 15-m radius, and for regeneration (11%) in a 10-m or 5-m whether the maximum DBH was greater than 12.5 cm or lower, respectively. The metrics were calculated using elevation and intensity values, excluding the echoes below 3 m since it was considered that those returns belonged to the bushy vegetation or trees in regeneration and were beyond the scope of this study. Afterward, the same canopy metrics were also calculated over the entire surface of Catalonia through a 20 m × 20 m regular grid.
We relied on the 5 m × 5 m Digital Terrain Model (DTM) elaborated by ICGC to derive a 20 m × 20 m regular grid model of the orthometric height, to then calculate the slope and aspect. The estimated altimetric accuracy was 0.90 m (RMSE).
Data preparation
A hundred LiDAR metrics were calculated for circular plots of 5-m, 10-m, 15-m, or 25-m radius, having the same center (the same x and y coordinates) as the NFI4 field plots. The initial LiDAR metrics included the percentiles of the height and intensity distributions of the echoes (1%, 5%, 10%, 20%, …, 90%, 95%, 99% percentiles), number and proportion of first echoes, minimum, mean, maximum, mode, variance, skewness, and kurtosis of the distributions of echo height and intensity, etc. The number of LiDAR metrics used in the analyses of this study was 53.
In addition to the LiDAR metrics, the x and y coordinates and a set of topographic variables were used as additional potential variables to be included in the distance measure. In Catalonia, the species composition changes rather systematically with the altitude. It can also be assumed that slope and aspect might be useful variables when finding the most similar forest inventory plot for a certain location. Slope correlates with soil depth which in turn correlates with site productivity. Aspect is correlated with moisture conditions and temperature, with northern aspects being moister and cooler than slopes facing to the south (Bonet et al. 2010).
The topographic variables calculated for each plot were altitude (m a.s.l.), slope (%) and aspect. As the significance of the aspect increases with increasing slope, the aspect was converted to two transformed variables, referred to as “northness” index (Eq. 1) and “eastness” index (Eq. 2) (Bonet et al. 2010, Fig. 2).
where, Aspect is the main compasses direction of the slope.
Optimizing the kNN imputation method
The purpose of the study was to find the optimal rule to select k nearest neighbors (kNN) for a certain forest inventory plot when LiDAR metrics, topographic variables, and x and y coordinates can be used to measure the distance between the plots. Weighted Euclidean distance was used as the distance measure (Eq. 3).
where, Dij is the distance between plots i and j, N is the number of variables included in the distance measure, wn is the weight of variable n, and xni and xnj are, respectively, the values of variable n in locations i and j. All variables were normalized using Eq. 4:
where, x is the normalized value, xOrig is the original value (in original units), xMin is the minimum value within the dataset in original units, and xMax is the maximum value in each dataset.
Each of the 59 variables (53 LiDAR metrics, 4 topographic variables, and x and y coordinates) could be used as such (non-transformed) or as transformed. The tested transformations were logarithm, square root, and square. Together with the transformations, the number of variables that could be used in the distance measure was 236.
Objective function
To measure the similarity of the measured and imputed tree lists, 14 variables were computed for each forest inventory plot and used to assess the similarity of the measured and imputed tree lists. These 14 variables included:
-
Total basal area, m2 ha−1
-
Total number of trees per hectare
-
Basal area of Pinus sylvestris, P. nigra, P. halepensis, P. uncinata, all oak species combined, and all other species combined (other than pines and oaks)
-
10%, 50% and 90% percentiles of the diameter distribution of the number of trees
-
10%, 50% and 90% percentiles of the diameter distribution of basal area
The minimized objective function in the optimization of the imputation rule was the weighted mean of the relative RMSEs of these 14 variables (Eq. 5).
where, RMSEi is the RMSE of variable i, \({\overline{x} }_{i}\) is the mean of variable i, and vi is the weight of variable i.
Most weight was given to the total basal area since it strongly correlates with the total volume, total biomass, carbon stock, and monetary value of the growing stock. It was also considered important that the imputed trees were like the measured trees in terms of the average tree size and species composition. Therefore, the 50% percentile of the basal area distribution (basal area median diameter) and basal area of the main tree species of the county were given much weight. The main tree species of Solsonès was P. nigra and the main species of Alt Urgell was P. sylvestris. As a result, the weight of the relative RMSE of the total basal area was 0.3226 (ten times higher than the lowest weight), the weights of the basal area of the main species and median diameter were 0.1613 (five times higher than the lowest weights), and all the other weights were 0.03226. The sum of the 14 weights was 1.
Optimization methods
The objective was to find the optimal measure to assess the similarity of two locations of a forested landscape, to be used in the imputation of forest data from one location to the other. The distance measure was based on Euclidean distance. Optimization consisted of selecting the best combination of variables among the 59 available variables as well as the optimal transformations and weights of these variables.
In the first variant of the optimization method (Fig. 3), simulated annealing (e.g., Bettinger et al. 2002) was used to select the optimal combination of variables and their transformations, using equal weights for all variables. In the second step, differential evolution (Storn and Price 1997) was employed to optimize the weights of the variables that had been selected for the distance measure. Simulated annealing (SA) was found to be the best or among the best metaheuristics to solve combinatorial optimization problems (Bettinger et al. 2002; Pukkala and Heinonen 2006; ** et al. 2016) and differential evolution (DE) has performed well in the optimization of continuous decision variables (Pukkala 2009; ** et al. 2018).
Flow chart of the two-step method for optimizing the distance measure in kNN data imputation. The first step finds the optimal combination of variables and their transformations when the weights of the variables are equal. The second step optimizes the weights of the variables selected in the first step
The number of variables used in the distance measure was set before the optimizations. Preliminary runs were done with 15 and 10 variables, and it was found that 10 variables gave equally good imputation results as the use of 15 variables. Packalen et al. (2012) found that even three LiDAR metrics may suffice in the distance measure when they are selected with simulated annealing. In our study, there was no need to try fewer than 10 variables because the optimization of variable weights could result in zero weights for some variables, which is equal to a priori reducing the number of variables used in the distance measure.
The SA heuristics was started by selecting a random combination of 10 variables, among the 59 available variables. Then, a random transformation was selected for each variable among the four alternatives (i.e., non-transformed, logarithm, square root, square). The Euclidean distance based on these variables and transformations was the initial solution of the SA run. This distance measure was used to find the k most similar plots for each NFI plot assuming that all wn in Eq. 3 were equal to one. These k plots were used to impute the 14 growing stock variables for the inventory plots. Inverted distances were used as the weights of the k nearest plots. Measured and imputed values of the 14 variables were used to calculate the objective function value (Eq. 5) for the initial solution.
Simulated annealing consists of making small changes in the initial solution and evaluating the effect of every change on the objective function. Changes are called moves, and each move produces a candidate solution. In this study, a move consisted of selecting one of the 10 variables included in the current distance measure and replacing this variable with another, randomly selected variable (from the list of 59 variables) and the corresponding randomly selected transformation. Because the new, randomly selected variable could be the same as the previous one, a move could be equal to selecting a different transformation of the same variable, the same transformation for a different variable, or a different transformation of a different variable.
All moves that improved the objective function (reduced the weighted mean RMSE of the 14 growing stock variables) were accepted and the other changes (inferior solutions) were accepted with probability.
where, T is the “temperature”, which affects the probability of accepting inferior solutions. The temperature is decreased during the optimization run, which means that the probability of accepting inferior solutions decreases along the optimization run. The purpose of accepting inferior solutions is to decrease the likelihood of getting trapped in a local optimum.
In the SA runs of this study, the initial temperature was set to 3. The number of candidates produced in each temperature was equal to the number of variables times the number of transformations (59 × 4 = 236). Then, the temperature was decreased by multiplying it by 0.95, and another 236 candidates were produced and evaluated. The process was terminated when the temperature reached a “freezing temperature”, which was equal to 0.01 times the initial temperature. Setting the starting temperature usually requires some testing and prior knowledge of the magnitude of the objective function. A suitable value for the starting temperature is about the same as the maximum effect of a move on the objective function value (Pukkala and Heinonen 2006).
The SA run produced the optimal set of ten variables and their optimal transformations for the distance measure, under the assumption that the weights of all variables were equal (Fig. 3). Differential evolution was used to examine if the distance measure could be improved by optimizing the weights of the variables (wn in Eq. 3). Differential evolution is a population-based optimization method for continuous variables, which means that the algorithm operates with several solutions that are modified and combined to obtain new candidate solutions (Storn and Price 1997; Pukkala 2009). A recommended population size is about ten times the number of optimized variables. Therefore, the population size used in this study was set to 100.
The initial solution vectors (vectors of variable weights wn in Eq. 3) were random numbers uniformly distributed between 0 and 1 which were subsequently scaled so that their mean was equal to one. The objective function value (Eq. 5) was calculated for every solution vector. Then, all the solution vectors were modified for several iterations, one solution vector at a time. In this process, the values of the elements of the solution vectors were either kept unchanged or picked from a “noise vector”, generated separately for each solution vector. The noise vector was produced from three other, randomly selected solution vectors as follows (Storn and Price 1997):
where, yi is element i of the noise vector, and xAi, xBi, and xCi are the values of the same element in three randomly selected solution vectors, and λ is a parameter (0.5 used in this study). The element was replaced by the noise vector value with a probability of 0.5. However, in one, randomly selected solution vector, every element was replaced by the noise vector value.
If the modified solution vector improved the objective function value, it replaced the previous vector. Otherwise, the solution vector was kept unchanged. This process of modifying and evaluating the solution vectors was repeated for 50 iterations, which was found sufficient in the optimization problem of the current study.
The two-step optimization process described above (using SA to select variables and their transformations, and subsequently using DE to optimize the weights of the selected variables) ignores the possibility that the optimal weights may change when the combination of the variables and transformations used in the distance measure is altered. Therefore, another variant for optimizing the distance measure was tested in which the weights were optimized simultaneously with the selection of variables and their transformations. However, to keep the computational burden bearable, the weights were discretized by allowing only three values (0.5—low, 1.0—average, and 1.5—high) while the number of transformations was reduced to three (non-transformed, square root, square) since logarithm multiplied by its optimal weight produces a similar relationship as square root multiplied by its optimal weight.
In this second variant of the optimization method, a candidate solution consisted of a set of 10 variables, their transformations, and weights. Since each variable had nine possible combinations of weight and transformation, the complexity of the problem was equal to selecting the best combination of 10 variables from a set of 531 variables (59 × 3 × 3 = 531). Only SA was applied in this optimization problem. In this case, a move consisted of selecting a random member of the 10 members of the current solution. Then, it was replaced by a new member, obtained by selecting, first, a random variable (from the list of 59 variables), second, a random transformation, and third, a random weight for the selected variable. Otherwise, the SA algorithm was the same as described above.
Results
Number of nearest neighbors
The optimization methods described above did not optimize the number of nearest neighbors that were used in data imputation. This question was analyzed by comparing the optimization results when the imputed values were based on k = 1, 3, 5, or 7 nearest neighbors. The inverted distance was used as the weight of the neighbors. The distance was calculated with Eq. 3.
The results showed that using only one neighbor was inferior to the use of 3, 5 or 7 nearest neighbors (Fig. 4). The results of Fig. 4 are based on the two-step method where the variables and their transformations were optimized in the first step (using SA), and variable weights were optimized in the second step (using DE). In Solsonès, the objective function value improved with increasing number of neighbors, but the differences between 3, 5, or 7 neighbors were small. In Alt Urgell, the use of 5 nearest neighbors minimized the weighted mean of the relative RMSEs of the 14 growing stock variables included in the objective function (Eq. 5).
Weighted mean of the relative RMSEs of the 14 growing stock variables included in the objective function when 1, 3, 5 or 7 nearest neighbors were used in data imputation (nn1, nn3, nn5, and nn7, respectively, on the x-axis). The results are averages of five repeated optimizations. Step 1 used simulated annealing to find the best combination of 10 variables and their optimal transformations for the distance measure. Step 2 optimized the weights of the selected variables applying differential evolution
Figure 4 also shows the improvement that was obtained by optimizing the weights of the variables of the distance function (wk in Eq. 3). The improvements were larger in Alt Urgell, as compared to Solsonès. The improvement obtained by weighting depended also on the number of neighbors used in data imputation.
Figure 5 shows the effect of the number of nearest neighbors and variable weighting on the RMSE of total basal area and basal area median diameter (50% percentile of the diameter distribution of basal area), and bias of total basal area. The relative RMSEs did not change much when the weights of the variables used in the distance measure were optimized (Fig. 5). However, optimizing the weights of the variables had a clear effect on the bias of total basal area (Fig. 5C). The effect of weight optimization was not the same in Solsonès and Alt Urgell (Fig. 5).
Relative RMSE of basal area (A) and basal area median diameter (B), and average absolute bias of basal area (C) when data imputation is based on 1 (nn1), 3 (nn3), 5 (nn5) or 7 (nn7), nearest neighbors. The results are averages of five repeated optimizations. Step 1 used simulated annealing to find the best combination of 10 variables and their optimal transformations for the distance measure. Step 2 optimized the weights of the selected variables using differential evolution
Comparison of alternative methods to optimize the distance measure
Based on the comparisons of objective function values (Fig. 4), RMSEs (Fig. 5A, B) and biases (Fig. 5C) it was concluded that using three to five nearest neighbors is sufficient when imputing forest inventory data for a certain geographical location in the Solsonès or Alt Urgell counties of Catalonia. The two different variants of the algorithm for optimizing the distance measure were compared in the case of three or five nearest neighbors.
Figure 6 shows that the best results were obtained for the two-step method in which the variables and their transformations were optimized in the first step, and variable weights were optimized in the second step. In Alt Urgell, the one-step method that included a rough optimization of variable weights (low/average/high) was better than the first step of the two-step method that assumed equal weights for all variables.
Average objective function value in five repeated optimizations of the distance measure after the first and second step of the two-step optimization method, and in the one-step method when 3 or 5 nearest neighbors are used in data imputation (nn3 and nn5, respectively, in the x-axis). Step 1 of the two-step method used simulated annealing to find the best combination of 10 variables and their optimal transformations for the distance measure. Step 2 optimized the weights of the selected variables applying differential evolution. The one-step method used simulated annealing to find the optimal combination of variables, and their transformations and weights
Overall, the best way to optimize the distance measure of kNN imputation consisted of selecting the optimal variables and transformations in the first step and optimizing the weights of the selected variables in the second step.
Variables selected for the distance measure
Altogether, 30 optimizations were conducted to produce the results shown in Figs. 4, 5 and 6. The variables most often selected for the distance measure were different for the two counties. Variables other than LiDAR metrics were frequently included in the distance measure (Table 1).
Location (x and y) or topographic variables were always selected for the distance measure. In Solsonès, the altitude of the terrain was always included in the distance measure, its most common transformation being square, followed by a non-transformed value. This implies that in this area, data for a certain location should be imputed from the same altitude. In Alt Urgell, the y coordinate was the most frequently used variable, suggesting that data should be imputed from the same latitude. All four transformations of the y coordinate were used. In Alt Urgell, altitude was included in 14 optimized distance measures, and the square was the optimal transformation of altitude in 7 out of 14 cases.
Because the distance measure included up to 10 variables, and among the 59 variables available there were only 6 that were not LiDAR metrics, several LiDAR metrics were selected in every distance measure. However, the combination of the selected LiDAR metrics varied from case to case. This is because many of the 53 LiDAR metrics correlated strongly with several other metrics. For example, at least one of the percentiles of the distribution of echo height was selected in every distance measure but the selected percentiles varied from case to case.
The weights of variables included in the distance measure also reflect their importance. In the first step of the two-step method, all weights were equal to 1. The second step optimized the weights by kee** their mean equal to 1. In Solsonès, optimization increased the weight of altitude, on average, from 1.00 to 1.35. In Alt Urgell, the average weight of the y coordinate increased from 1.00 to 1.84. The results indicate that the variables that were most often included in the distance measure also had a higher weight.
Optimal distance measure
The distance measure that minimized the weighted mean of the relative RMSEs of the 14 growing stock variables included the variables shown in Table 2. The lists included nine and eight variables for Alt Urgell and Solsonès, respectively, although ten variables were selected in the first step of the optimization process. However, in the second step, the optimized weight was zero for one and two variables in Alt Urgell and Solsonès, respectively.
It may be assumed that the variables selected for the distance measure correlate with the growing stock variables. Most probably, altitude and latitude (y coordinate) are useful for selecting neighbors that have the same species composition as the location to which data are imputed. Figure 7 shows that some of the percentiles of the distribution of echo heights correlated with the stand basal area. In addition, skewness of the distribution of echo heights correlated strongly with basal area median diameter. However, several of the variables selected for the distance measure showed no clear correlation with the growing stock variables.
Correlation of three LiDAR metrics included in the best distance measures with stand basal area or basal area median diameter. Lidar15 is the skewness of the distribution of echo heights, Lidar21 is the 20% percentile of the distribution of echo heights, and Lidar32 is the 99% percentile of the distribution of echo heights
Imputation example
The Solsonès and Alt Urgell counties were scanned completely, and the same LiDAR metrics that were tested for the distance measure were also calculated for 20 m × 20 m grid cells. Therefore, the forest inventory data can be imputed for any location of these counties, using the distance measures developed in this study. Figure 8 shows some imputation results for a randomly selected 2 km × 2 km grid in Alt Urgell.
Discussion
This study developed a ready-to-use methodology for obtaining wall-to-wall forest inventory data for any forested area of Catalonia. The methodology refers to the optimization of the distance measure and the subsequent use of this measure in actual data imputation. Our analyses led to the same conclusion as done by Chirici et al. (2008) and Packalen et al. (2012), namely that it is not possible to develop a single distance measure that is valid for different biogeographical areas. Instead, data imputation should be started by optimizing the distance measure. Compared to Packalen et al. (2012), the novelty of our method was that the transformations of the variables included in the distance measure were also optimized, in addition to the combination of them, and the weights of the variables.
However, not all analyses conducted in this study need to be repeated when applying the methodology to different geographical areas, as we showed that 3NN, i.e., using three nearest neighbors, was appropriate and the two-step optimization method worked best. Our results differ from those of LeMay and Temesgen (2005) who did not obtain large gains in using the average of three neighbors rather than a single neighbor.
Our study also showed that it was worthwhile to include topographic variables, as well as x and y coordinates, and not only LiDAR metrics, in the set of variables from which the optimal combination is selected. The obvious reason for the importance of these variables is the natural gradual transitions in the species composition and growing stock density of Mediterranean forests, transitions that operate along a north–south topographic-climatic gradient (Scarascia-Mugnozza et al. 2000, Vilà-Cabrera et al. 2011). Compared to some other European countries, natural transitions in forest featured are less disrupted by management (Palahí et al. 2008). As most trees in Catalan forests are pine species, that might be difficult to differentiate from each other from LiDAR data, variables other than LiDAR metrics are useful to find neighbors that are similar in terms of species composition.
The analyses conducted for the two counties (Solsonès and Alt Urgell) showed that quite different distance measures were optimal in these counties. The most important single variable seemed to be altitude in Solsonès and y coordinate (latitude) in Alt Urgell. In addition, the effect of optimizing the weights of the variables was larger in Alt Urgell than in Solsonès. The results also suggested that ten variables are enough to be included in the distance measure. Indeed, the optimal weight of the selected variables was sometimes zero, meaning that there was no need to include more than ten variables in the optimized distance measures.
When values of the growing stock variables imputed to the NFI plots were compared to the measured values, the RMSE was around 22% for basal area median diameter and 35–40% for the total basal area. The RMSE was around 20% for the 90% percentile of the diameter distribution. The relative RMSEs were higher for species-specific basal areas than for total basal area. In comparison, in Packalen et al. (2012) the RMSE was less than 10% for dominant height when it was the only imputed variable. When five variables were imputed simultaneously, relative RMSEs of 48–80% were obtained for species-specific volumes, number of trees per hectare, and basal area median diameter. Chirici et al. (2008) reached a relative RMSE of 44–63% in the volume estimation for their Mediterranean research area. In terms of the RMSEs of growing stock variables, our results are comparable to those of Packalen et al. (2012). The bias of the imputed stand basal area was usually below 5% of the average basal area of the NFI plots when 3 or 5 neighbors were used in imputation. Both studies suggest that the size of the largest trees is the most accurately imputed information, most probably because dominant height correlates strongly with the percentiles of echo heights, and tree diameter correlates strongly with tree height.
However, the imputed values still differed a lot from the values of growing stock variables calculated from the trees measured in the NFI plots. The magnitude of the RMSE of the stand basal area was 9 m2 ha−1. It means that in about one-third of the plots, the measured and imputed basal areas differed more than 9 m2 ha−1. A possible reason for that is the fact that the Spanish NFI uses concentric plots as a sampling design. As a result, the trees were not measured from exactly the area for which the LiDAR metrics were calculated. For example, trees larger than 42.5 cm in DBH were measured within the 25-m radius sub-plot and trees between 7.5 and 12.5 cm in DBH were measured within the 10-m radius sub-plot. However, the LiDAR metrics were mostly computed for a circular area with a 15- or 20-m radius. Another drawback in the NFI data was that small trees DBH (< 7.5 cm) were not sampled.
One reason for decreased RMSEs was that good results for 14 variables were pursued simultaneously. Indeed, the results would have been better when only one variable was imputed (Packalen et al. 2012).
Another possible reason behind the differences between growing stock variables derived from imputed and sampled tree lists is that the LiDARCAT2 flights that covered the two counties were run during different seasons and years (Fig. 1). Analysis based on LiDAR data collected during spring or summer and winter without differentiating them may entail mismatches, especially in deciduous forests.
Currently, the same LiDAR metrics and topographic variables that were used in this study are calculated for 20 m × 20 m raster cells over the whole of Catalonia. Therefore, the logical first step is to impute forest data for these cells. In our study, forest data refers to the list of trees measured in NFI plots. The most succinct way to store the imputation result consists of the numbers of the three nearest NFI plots and their weights (inverted distances). Applications can then retrieve these NFI plots and generate a tree list for the raster cells. If the calculation unit is a stand or segment, the tree lists can be produced by merging the tree lists of the cells that constitute these larger spatial units. Optimizing the distance measure for the different biogeographical regions of Catalonia and applying it to 20 m × 20 m raster cells would also allow the production of a continuous forest map for the entire Catalonia.
The imputation results based on an optimized similarity model can become outdated in a relatively short time. To overcome this limitation, it is necessary to update the forest imputation results whenever new LiDAR or forest inventory data are available. Indeed, the LiDARCAT3 project to cover all of Catalonia is currently on a mission with the sensors Hyperson2+ and MFC150 for the LiDAR and optical images, respectively, to get 10 points m−2 coverture. Once the same set of derived metrics from the cloud point is released, applying the same distance metric will provide updated imputation results.
Conclusion
The results of the study indicated that topographic variables and latitude are important variables, in addition to LiDAR metrics, in the kNN forest data imputation in Catalonia. The study also showed that the optimal imputation method may be different for different geographical areas, even in different municipalities of the same province of the same country. Therefore, the first step of kNN data imputation should always optimize the imputation method. This study developed a methodology for optimized kNN data imputation for Catalonian forests. In this method, forest data are imputed for grids of raster cells from the field plots of the national forest inventory.
References
Alberdi I, Sandoval V, Condes S, Cañellas I, Vallejo R (2016) El Inventario Forestal Nacional español, una herramienta para el conocimiento, la gestión y la conservación de los ecosistemas forestales arbolados. Ecosistemas 25(3):88–97. https://doi.org/10.7818/ECOS.2016.25-3.10
Bettinger P, Graetz D, Boston K, Sessions J, Chung W (2002) Eight heuristic planning techniques applied to three increasingly difficult wildlife planning problems. Silva Fennica 36(2):561–584
Blázquez-Casado Á, González-Olabarria JR, Martín-Alcón S, Just A, Cabré M, Coll L (2015) Assessing post-storm forest dynamics in the Pyrenees using high-resolution LIDAR data and aerial photographs. J Mt Sci 12:841–853. https://doi.org/10.1007/s11629-014-3327-3
Bonet JA, Palahí M, Colinas C, Pukkala T, Fischer C, Miina J, Martinez de Aragón J (2010) Modelling the production of wild mushrooms in pine forests in the Central Pyrenees in northeastern Spain. Can J for Res 40:347–356. https://doi.org/10.1139/X09-198
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/A:1010933404324
Chirici G, Barbati A, Corona P, Marchetti M, Travaglini D, Maselli F, Bertini R (2008) Non-parametric and parametric methods using satellite images for estimating growing stock volume in alpine and Mediterranean forest ecosystems. Remote Sens Environ 112(5):2686–2700. https://doi.org/10.1016/j.rse.2008.01.002
Crookston NL, Finley A (2008) yaImpute: an R Package for kNN imputation. J Stat Softw 23(10). Available on http://www.jstatsoft.org/
Díaz-Yáñez O, Pukkala T, Packalen P, Peltola H (2020) Multifunctional comparison of different management strategies in boreal forests. Forestry 93(1):84–95. https://doi.org/10.1093/forestry/cpz053
FUSION, version 3.2 (2012) – LiDAR analysis and visualization software. Available on: http://forsys.sefs.uw.edu/fusion/fusion_overview.html. Accessed 18 May 2023
Gittins R (1985) Canonical analysis: a review with applications in ecology. Springer-Verlag, Berlin. p, p 351
Hudak AT, Crookston NL, Evans JS, Hall DE, Falkowski MJ (2008) Nearest neighbor imputation of species-level, plot-scale forest structure attributes from LiDAR data. Remote Sens Environ 112(5):2232–2245. https://doi.org/10.1016/j.rse.2007.10.009
Hyyppä J, Hyyppä H, Leckie D, Gougeon F, Yu X, Maltamo M (2008) Review of methods of small-footprint airborne laser scanning for extracting forest inventory data in boreal forests. Int J Remote Sens 29(5):1339–1366. https://doi.org/10.1080/01431160701736489
Jia W, Sun Y, Pukkala T, ** X (2020) Improved cellular automaton for stand delineation. Forests 11(1):37. https://doi.org/10.3390/f11010037
** X, Pukkala T, Li F (2016) Fine-tuning heuristic methods for combinatorial optimization in forest planning. Eur J Forest Res 135:765–779. https://doi.org/10.1007/s10342-016-0971-x
** X, Pukkala T, Li F (2018) Meta optimization of stand management with population-based methods. Can J for Res 48:697–708. https://doi.org/10.1139/cjfr-2017-0404
Latifi H, Nothdurft A, Koch B (2010) Non-parametric prediction and map** of standing timber volume and biomass in a temperate forest: application of multiple optical/LiDAR-derived predictors. Forestry 83(4):395–407. https://doi.org/10.1093/forestry/cpq022
LeMay V, Temesgen H (2005) Comparison of nearest neighbor methods for estimating basal area and stems per hectare using aerial auxiliary variables. Forest Sci 51(2):109–119
Lim K, Treitz P, Wulder M, St-Onge B, Flood M (2003) LiDAR remote sensing of forest structure. Prog Phys Geogr Earth Environ 27(1):88–106. https://doi.org/10.1191/0309133303pp360ra
Maltamo M, Malinen J, Packalén P, Suvanto A, Kangas J (2006) Nonparametric estimation of stem volume using airborne laser scanning, aerial photography, and stand-register data. Can J For Res 36:426–436. https://doi.org/10.1139/x05-246
Martín-Alcón S, Coll L, De Cáceres M, Guitart L, Cabré M, Just A, González-Olabarria JR (2015) Combining aerial LiDAR and multispectral imagery to assess post-fire regeneration types in a Mediterranean forest. Can J For Res 45(7):56866. https://doi.org/10.1139/cjfr-2014-0430
Moeur M, Stage AR (1995) Most similar neighbor: an improved sampling inference procedure for natural resource planning. Forest Sci 41(2):337–359. https://doi.org/10.1093/forestscience/41.2.337
Packalen P, Temesgen H, Maltamo M (2012) Variable selection strategies for nearest neighbor imputation methods used in remote sensing based forest inventory. Can J Remote Sens 38(5):557–569. https://doi.org/10.5589/m12-046
Palahí M, Mavsar R, Gracia C, Birot Y (2008) Mediterranean forests under focus. Int Forest Rev 10(4):676–688. https://doi.org/10.1505/ifor.10.4.676
Pukkala T (2009) Population-based methods in the optimization of stand management. Silva Fennica 43(2):261–274. https://doi.org/10.14214/sf.211
Pukkala T (2019) Using ALS raster data in forest planning. J Forest Res 30:1581–1593. https://doi.org/10.1007/s11676-019-00937-6
Pukkala T (2020) Delineating forest stands from grid data. Forest Ecosyst 7:1–14. https://doi.org/10.1186/s40663-020-00221-8
Pukkala T, Heinonen T (2006) Optimizing heuristic search in forest planning. Nonlinear Anal Real World Appl 7(5):1284–1297. https://doi.org/10.1016/j.nonrwa.2005.11.011
Rouget M, Richardson DM, Lavorel S, Vayreda J, Gracia C, Milton SJ (2001) Determinants of distribution of six Pinus species in Catalonia. Spain J Veg Sci 12(4):491–502. https://doi.org/10.2307/3237001
Scarascia-Mugnozza G, Oswald H, Piussi P, Radoglou K (2000) Forests of Mediterranean region: gaps in knowledge and research needs. For Ecol Manage 132:97–109. https://doi.org/10.1016/S0378-1127(00)00383-2
Storn R, Price K (1997) Differential evolution – a simple and efficient heuristic for global optimization over continuous spaces. J Global Optim 11:341–359. https://doi.org/10.1023/A:1008202821328
Terrasolid version 017 (2017) – The standard workflow for airborne LiDAR classification. Available on: https://terrasolid.com/. Accessed on 17 May 2023
Trasobares A, Mola-Yudego B, Aquilué N, González-Olabarria JR, Garcia-Gonzalo J, García-Valdés R, De Cáceres M (2022) Nationwide climate-sensitive models for stand dynamics and forest scenario simulation. For Ecol Manage 505:119909. https://doi.org/10.1016/j.foreco.2021.119909
Vilà-Cabrera A, Martínez-Vilalta J, Vayreda J, Retana J (2011) Structural and climatic determinants of demographic rates of Scots pine forests across the Iberian Peninsula. Ecol Appl 21:1162–1172. https://www.jstor.org/stable/23022987
White JC, Wulder MA, Varhola A, Vastaranta M, Coops NC, Cook BD, Pitt D, Woods M (2013) A best practices guide for generating forest inventory attributes from airborne laser scanning data using an area-based approach. Canadian Forest Service Canadian Wood Fibre Centre Information Report FI-X-010
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This work was supported by a Juan de la Cierva fellowship of the Spanish Ministry of Science and Innovation (FCJ2020-046387-I) and the Spanish Ministry of Science, Innovation and Universities (PID2020-120355RB-IOO).
The online version is available at https://springer.longhoe.net/.
Corresponding editor: Yu Lei
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pukkala, T., Aquilué, N., Just, A. et al. Develo** kNN forest data imputation for Catalonia. J. For. Res. 35, 80 (2024). https://doi.org/10.1007/s11676-024-01735-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11676-024-01735-5