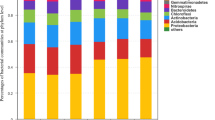
Abstract
Background and aims
Afforestation can alter belowground microbial diversity and affect soil‒plant feedback, which is crucial for understanding nutrient cycles and ecosystem productivity. Most studies have focused on the carbon and nitrogen cycles in forest ecosystems; however, there are few studies on the effect of afforestation in soil microbial diversity on phosphorus (P) nutrients. In this study, we explored the effect of soil microbial communities on soil‒plant P feedback under long-term afforestation.
Methods
Typical poplar plantations were selected from the Lhasa River Basin. We assessed the P content, activity of extracellular enzyme related to P acquisition, rhizosphere soil microbes, and mechanism of microbial community.
Results
Increased bacterial richness and evenness were beneficial to soil and plant P nutrition; however, the community differentiation of bacteria and fungi had no positive effect on the P content. Ecological stochasticity drove for the assembly of the rhizosphere soil microbial community after afforestation on the Tibetan Plateau. Homogenizing dispersal dominated bacterial and fungal assemblies. Ecological drift was another stochastic process that affected the microbial assembly.
Conclusion
Bacterial richness and evenness had positive effects on soil‒plant P feedback, whereas microbial differentiation did not. Homogenizing dispersal and ecological drift are intrinsic factors that regulate the effects of microbial community diversity on soil‒plant P feedback processes.
This study provides the evidence on the intrinsic mechanism of belowground microbial diversity influencing soil‒plant P feedback. It also provides new insights into the selection of plantation species on the Tibetan Plateau from the perspective of enhancement of P nutrition by microbes.







Similar content being viewed by others
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Chen et al. 2021) in National Genomics Data Center (CNCB-NGDC Members and Partners 2022), China National Center for Bioinformation / Bei**g Institute of Genomics, Chinese Academy of Sciences (GSA: CRA006086 for 16S rRNA gene amplicon datasets and CRA006090 for ITS2 gene amplicon datasets) that would be publicly accessible at https://ngdc.cncb.ac.cn/gsa. The data for soil P, SP, soil pH, root P, branch P, and leaf P can be found in Table S1.
Abbreviations
- AIC:
-
Akaike information criterion
- βMNTD:
-
The beta mean nearest taxon distance
- βNTI:
-
The beta nearest taxon index
- C:
-
Carbon
- Df:
-
Degree of freedom
- LDA:
-
Linear discriminant analysis
- LEfSe:
-
LDA effect size
- MCD:
-
Microbial community structure diversity
- NMDS:
-
Nonmetric multidimensional scaling
- OTUs:
-
Operational Taxonomic Units
- P:
-
Phosphorus
- PA:
-
Populus alba L.
- PB:
-
Populus × bei**gensis
- PC:
-
The blank plots
- Po:
-
Organic phosphorus
- PSM:
-
Phosphate solubilizing microbes
- PZ:
-
Populus szechuanica Var. tibetica Schneid.
- R2 :
-
The amount of variance explained by the model
- Rc 2 :
-
The conditional R2
- Rm 2 :
-
The marginal R2
- RCBray :
-
The taxonomic beta diversity metric Bray–Curtis-based Raup–Crick
- SEM:
-
The structural equation model
- SP:
-
Soil phosphatase
- VIFs:
-
Variance inflation factors
References
Allison SD, Hanson CA, Treseder KK (2007) Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol Biochem 39:1878–1887. https://doi.org/10.1016/j.soilbio.2007.02.001
Aßhauer KP, Wemheuer B, Daniel R, Meinicke P (2015) Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31:2882–2884. https://doi.org/10.1093/bioinformatics/btv287
Bahram M, Kohout P, Anslan S, Harend H, Abarenkov K, Tedersoo L (2016) Stochastic distribution of small soil eukaryotes resulting from high dispersal and drift in a local environment. ISME J 10:885–896. https://doi.org/10.1038/ismej.2015.164
Baldrian P (2017) Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol Rev 41:109–130. https://doi.org/10.1093/femsre/fuw040
Battini F, Grønlund M, Agnolucci M, Giovannetti M, Jakobsen I (2017) Facilitation of phosphorus uptake in maize plants by mycorrhizosphere bacteria. Sci Rep 7:4686. https://doi.org/10.1038/s41598-017-04959-0
Bergkemper F, Schöler A, Engel M, Lang F, Krüger J, Schloter M, Schulz S (2016) Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ Microbiol 18:1988–2000. https://doi.org/10.1111/1462-2920.13188
Bever JD, Dickie IA, Facelli E, Facelli JM, Klironomos J, Moora M, Rillig MC, Stock WD, Tibbett M, Zobel M (2010) Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol 25:468–478. https://doi.org/10.1016/j.tree.2010.05.004
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. https://doi.org/10.1038/nmeth.2276
Bolan NS (1991) A critical review on the role of mycorrhizal fungi in the uptake of phosphorus by plants. Plant Soil 134:189–207. https://doi.org/10.1007/BF00012037
Bulgarelli D, Schlaeppi K, Spaepen S, Van Themaat EVL, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. https://doi.org/10.1146/annurev-arplant-050312-120106
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Chen T, Chen X, Zhang S, Zhu J, Tang B, Wang A, Dong L, Zhang Z, Yu C, Sun Y, Chi L, Chen H, Zhai S, Sun Y, Lan L, Zhang X, **ao J, Bao Y, Wang Y, Zhang Z, Zhao W (2021) The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics 19:578–583. https://doi.org/10.1016/j.gpb.2021.08.001
Chesson P (2000) Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst 31:343–366. https://www.jstor.org/stable/221736
Cleveland CC, Houlton BZ, Smith WK, Marklein AR, Reed SC, Parton W, Del Grosso SJ, Running SW (2013) Patterns of new versus recycled primary production in the terrestrial biosphere. Proc Natl Acad Sci U S A 110:12733–12737. https://doi.org/10.1073/pnas.1302768110
CNCB-NGDC Members and Partners (2022) Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res 50:D27–D38. https://doi.org/10.1093/nar/gkab951
de-Bashan LE, Magallon-Servin P, Lopez BR, Nannipieri P (2022) Biological activities affect the dynamic of P in dryland soils. Biol Fertil Soils 58:105–119. https://doi.org/10.1007/s00374-021-01609-6
Declerck SA, Winter C, Shurin JB, Suttle CA, Matthews B (2013) Effects of patch connectivity and heterogeneity on metacommunity structure of planktonic bacteria and viruses. ISME J 7:533–542. https://doi.org/10.1038/ismej.2012.138
Delgado-Baquerizo M, Giaramida L, Reich PB, Khachane AN, Hamonts K, Edwards C, Lawton LA, Singh BK (2016a) Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J Ecol 104:936–946. https://doi.org/10.1111/1365-2745.12585
Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, Berdugo M, Campbell CD, Singh BK (2016b) Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun 7:10541. https://doi.org/10.1038/ncomms10541
Delgado-Baquerizo M, Eldridge DJ, Ochoa V, Gozalo B, Singh BK, Maestre FT (2017) Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol Lett 20:1295–1305. https://doi.org/10.1111/ele.12826
De Mendiburu F (2021) agricolae: Statistical Procedures for Agricultural Research. R package version 1.3–5. https://CRAN.R-project.org/package=agricolae
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Fargione J, Brown CS, Tilman D (2003) Community assembly and invasion: an experimental test of neutral versus niche processes. Proc Natl Acad Sci U S A 100:8916–8920. https://doi.org/10.1073/pnas.103310710
Fatemi FR, Fernandez IJ, Simon KS, Dail DB (2016) Nitrogen and phosphorus regulation of soil enzyme activities in acid forest soils. Soil Biol Biochem 98:171–179. https://doi.org/10.1016/j.soilbio.2016.02.017
Faucon MP, Houben D, Reynoird JP, Mercadal-Dulaurent AM, Armand R, Lambers H (2015) Advances and perspectives to improve the phosphorus availability in crop** systems for agroecological phosphorus management. Adv Agron 134:51–79. https://doi.org/10.1016/bs.agron.2015.06.003
Feng Y, Chen R, Stegen JC, Guo Z, Zhang J, Li Z, Lin X (2018) Two key features influencing community assembly processes at regional scale: initial state and degree of change in environmental conditions. Mol Ecol 27:5238–5251. https://doi.org/10.1111/mec.14914
Filippelli GM (2008) The global phosphorus cycle: past, present, and future. Elements 4:89–95. https://doi.org/10.2113/GSELEMENTS.4.2.89
Fodelianakis S, Lorz A, Valenzuela-Cuevas A, Barozzi A, Booth JM, Daffonchio D (2019) Dispersal homogenizes communities via immigration even at low rates in a simplified synthetic bacterial metacommunity. Nat Commun 10:1314. https://doi.org/10.1038/s41467-019-09306-7
Friesen ML, Porter SS, Stark SC, von Wettberg EJ, Sachs JL, Martinez-Romero E (2011) Microbially mediated plant functional traits. Ann Rev Ecol Evol Syst 42:23–46. https://doi.org/10.1146/annurev-ecolsys-102710-145039
Gibson T (1988) Carbohydrate metabolism and phosphorus/salinity interactions in wheat (Triticum aestivum L.). Plant Soil 111:25–35. https://doi.org/10.1007/BF02182033
Giles CD, Richardson AE, Cade-Menun BJ, Mezeli MM, Brown LK, Menezes-Blackburn D, Darch T, Blackwell MSA, Shand CA, Stutter MI, Wendler R, Cooper P, Lumsdon DG, Wearing C, Zhang H, Haygarth PM, George TS (2018) Phosphorus acquisition by citrate-and phytase-exuding Nicotiana tabacum plant mixtures depends on soil phosphorus availability and root intermingling. Physiol Plant 163:356–371. https://doi.org/10.1111/ppl.12718
Hacker N, Ebeling A, Gessler A, Gleixner G, González Macé O, de Kroon H, Lange M, Mommer L, Eisenhauer N, Ravenek J, Scheu S, Weigelt A, Wagg C, Wilcke W, Oelmann Y (2015) Plant diversity shapes microbe-rhizosphere effects on P mobilisation from organic matter in soil. Ecol Lett 18:1356–1365. https://doi.org/10.1111/ele.12530
He Q, Wu Y, Bing H, Zhou J, Wang J (2020) Vegetation type rather than climate modulates the variation in soil enzyme activities and stoichiometry in subalpine forests in the eastern Tibetan Plateau. Geoderma 374:114424. https://doi.org/10.1016/j.geoderma.2020.114424
Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, Wu HC, Lara M, Town CD, Kopka J, Udvardi MK, Vance CP (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144:752–767. https://doi.org/10.1104/pp.107.096958
Huang CY, Roessner U, Eickmeier I, Genc Y, Callahan DL, Shirley N, Langridge P, Bacic A (2008) Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant Cell Physiol 49:691–703. https://doi.org/10.1093/pcp/pcn044
Izzo A, Agbowo J, Bruns TD (2005) Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. New Phytol 166:619–630. https://doi.org/10.1111/j.1469-8137.2005.01354.x
Jangid K, Whitman WB, Condron LM, Turner BL, Williams MA (2013) Soil bacterial community succession during long-term ecosystem development. Mol Ecol 22:3415–3424. https://doi.org/10.1111/mec.12325
Jiao S, Yang Y, Xu Y, Zhang J, Lu Y (2020) Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J 14:202–216. https://doi.org/10.1038/s41396-019-0522-9
**g X, Sanders NJ, Shi Y, Chu H, Classen AT, Zhao K, Chen L, Shi Y, Jiang Y, He JS (2015) The links between ecosystem multifunctionality and above-and belowground biodiversity are mediated by climate. Nat Commun 6:8159. https://doi.org/10.1038/ncomms9159
Karhu K, Wall A, Vanhala P, Liski J, Esala M, Regina K (2011) Effects of afforestation and deforestation on boreal soil carbon stocks—Comparison of measured C stocks with Yasso07 model results. Geoderma 164:33–45. https://doi.org/10.1016/j.geoderma.2011.05.008
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. https://doi.org/10.1093/bioinformatics/btq166
Knelman JE, Graham EB, Trahan NA, Schmidt SK, Nemergut DR (2015) Fire severity shapes plant colonization effects on bacterial community structure, microbial biomass, and soil enzyme activity in secondary succession of a burned forest. Soil Biol Biochem 90:161–168. https://doi.org/10.1016/j.soilbio.2015.08.004
Knelman JE, Legg TM, O’Neill SP, Washenberger CL, González A, Cleveland CC, Nemergut DR (2012) Bacterial community structure and function change in association with colonizer plants during early primary succession in a glacier forefield. Soil Biol Biochem 46:172–180. https://doi.org/10.1016/j.soilbio.2011.12.001
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5527. https://doi.org/10.1111/mec.12481
Koukol O, Novák F, Hrabal R, Vosátka M (2006) Saprotrophic fungi transform organic phosphorus from spruce needle litter. Soil Biol Biochem 38:3372–3379. https://doi.org/10.1016/j.soilbio.2006.05.007
Kulmatiski A, Beard KH, Stevens JR, Cobbold SM (2008) Plant–soil feedbacks: a meta-analytical review. Ecol Lett 11:980–992. https://doi.org/10.1111/j.1461-0248.2008.01209.x
Laliberté E, Grace JB, Huston MA, Lambers H, Teste FP, Turner BL, Wardle DA (2013) How does pedogenesis drive plant diversity? Trends Ecol Evol 28:331–340. https://doi.org/10.1016/j.tree.2013.02.008
Langenheder S, Székely AJ (2011) Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5:1086–1094. https://doi.org/10.1038/ismej.2010.207
Lefcheck JS (2016) piecewiseSEM: Piecewise structural equation modelling in r for ecology, evolution, and systematics. Methods Ecol Evol 7:573–579. https://doi.org/10.1111/2041-210X.12512
Li M, Wei Z, Wang J, Jousset A, Friman VP, Xu Y, Shen Q, Pommier T (2019a) Facilitation promotes invasions in plant-associated microbial communities. Ecol Lett 22:149–158. https://doi.org/10.1111/ele.13177
Li N, Dong K, Jiang G, Tang J, Xu Q, Li X, Kang Z, Zou S, Chen X, Adams JM, Zhao H (2020) Stochastic processes dominate marine free-living Vibrio community assembly in a subtropical gulf. FEMS Microbiol Ecol 96:fiaa198. https://doi.org/10.1093/femsec/fiaa198
Li X, Li Y, Peng S, Chen Y, Cao Y (2019b) Changes in soil phosphorus and its influencing factors following afforestation in Northern China. Land Degrad Dev 30:1655–1666. https://doi.org/10.1002/ldr.3345
Liu F, Chen L, Zhang B, Wang G, Qin S, Yang Y (2018) Ultraviolet radiation rather than inorganic nitrogen increases dissolved organic carbon biodegradability in a typical thermo-erosion gully on the Tibetan Plateau. Sci Total Environ 627:1276–1284. https://doi.org/10.1016/j.scitotenv.2018.01.275
Liu R, Yao Y, Li Q, Cai Z, Wei D, Wang X, Zhang S (2023) Rhizosphere soil microbes benefit carbon and nitrogen sinks under long-term afforestation on the Tibetan Plateau. Catena 220:106705. https://doi.org/10.1016/j.catena.2022.106705
Liu W, Liu L, Yang X, Deng M, Wang Z, Wang P, Yang S, Li P, Peng Z, Yang L, Jiang L (2021) Long-term nitrogen input alters plant and soil bacterial, but not fungal beta diversity in a semiarid grassland. Glob Chang Biol 27:3939–3950. https://doi.org/10.1111/gcb.15681
Lüdecke D, Ben-Shachar MS, Patil I, Waggoner P, Makowski D (2021) performance: An R package for assessment, comparison and testing of statistical models. J Open Source Softw 6:3139. https://doi.org/10.21105/joss.03139
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Mangan SA, Schnitzer SA, Herre EA, Mack KML, Valencia MC, Sanchez EI, Bever JD (2010) Negative plant–soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466:752–755. https://doi.org/10.1038/nature09273
Maynard DS, Crowther TW, Bradford MA (2017) Competitive network determines the direction of the diversity–function relationship. Proc Natl Acad Sci U S A 114:11464–11469. https://doi.org/10.1073/pnas.1712211114
Miki T, Yokokawa T, Matsui K (2014) Biodiversity and multifunctionality in a microbial community: a novel theoretical approach to quantify functional redundancy. Proc R Soc B 281:20132498. https://doi.org/10.1098/rspb.2013.2498
Monard C, Mchergui C, Nunan N, Martin-Laurent F, Vieuble-Gonod L (2012) Impact of soil matric potential on the fine-scale spatial distribution and activity of specific microbial degrader communities. FEMS Microbiol Ecol 81:673–683. https://doi.org/10.1111/j.1574-6941.2012.01398.x
Mouquet N, Loreau M (2003) Community patterns in source-sink metacommunities. Am Nat 162:544–557. https://doi.org/10.1086/378857
Nacke H, Goldmann K, Schöning I, Pfeiffer B, Kaiser K, Castillo-Villamizar GA, Schrumpf M, Buscot F, Daniel R, Wubet T (2016) Fine spatial scale variation of soil microbial communities under European beech and Norway spruce. Front Microbiol 7:2067. https://doi.org/10.3389/fmicb.2016.02067
Neal AL, Rossmann M, Brearley C, Akkari E, Guyomar C, Clark IM, Allen E, Hirsch PR (2017) Land-use influences phosphatase gene microdiversity in soils. Environ Microbiol 19:2740–2753. https://doi.org/10.1111/1462-2920.13778
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2020) vegan: Community Ecology Package. R package version 2.5–7
Osorio NW, Habte M (2014) Soil phosphate desorption induced by a phosphate-solubilizing fungus. Commun Soil Sci Plant Anal 45:451–460. https://doi.org/10.1080/00103624.2013.870190
Peng Z, Wang Z, Liu Y, Yang T, Chen W, Wei G, Jiao S (2021) Soil phosphorus determines the distinct assembly strategies for abundant and rare bacterial communities during successional reforestation. Soil Ecol Lett 3:342–355. https://doi.org/10.1007/s42832-021-0109-z
Piao S, Huang M, Liu Z, Wang X, Ciais P, Canadell JG, Wang K, Bastos A, Friedlingstein P, Houghton RA, Quéré CL, Liu Y, Myneni RB, Peng S, Pongratz J, Sitch S, Yan T, Wang Y, Zhu Z, Wu D, Wang T (2018) Lower land-use emissions responsible for increased net land carbon sink during the slow warming period. Nature Geosci 11:739–743. https://doi.org/10.1038/s41561-018-0204-7
Pinheiro J, Bates D, R Core Team (2022) nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1–157, https://CRAN.R-project.org/package=nlme
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Ren C, Zhou Z, Guo Y, Yang G, Zhao F, Wei G, Han X, Feng L, Feng Y, Ren G (2021) Contrasting patterns of microbial community and enzyme activity between rhizosphere and bulk soil along an elevation gradient. Catena 196:104921. https://doi.org/10.1016/j.catena.2020.104921
Revillini D, Gehring CA, Johnson NC (2016) The role of locally adapted mycorrhizas and rhizobacteria in plant–soil feedback systems. Funct Ecol 30:1086–1098. https://doi.org/10.1111/1365-2435.12668
Reynolds HL, Packer A, Bever JD, Clay K (2003) Grassroots ecology: plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology 84:2281–2291. https://doi.org/10.1890/02-0298
Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C (2009) Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321:305–339. https://doi.org/10.1007/s11104-009-9895-2
Richardson AE, Simpson RJ (2011) Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol 156:989–996. https://doi.org/10.1104/pp.111.175448
Sánchez-Marañón M, Miralles I, Aguirre-Garrido JF, Anguita-Maeso M, Millán V, Ortega R, García-Salcedo JA, Martínez-Abarca F, Soriano M (2017) Changes in the soil bacterial community along a pedogenic gradient. Sci Rep 7:14593. https://doi.org/10.1038/s41598-017-15133-x
Seeling B, Zasoski RJ (1993) Microbial effects in maintaining organic and inorganic solution phosphorus concentrations in a grassland topsoil. Plant Soil 148:277–284. https://doi.org/10.1007/BF00012865
Semchenko M, Leff JW, Lozano YM, Saar S, Davison J, Wilkinson A, Jackson BG, Pritchard WJ, De Long JR, Oakley S, Mason KE, Ostle NJ, Baggs EM, Johnson D, Fierer N, Bardgett RD (2018) Fungal diversity regulates plant-soil feedbacks in temperate grassland. Sci Adv 4:eaau4578. https://doi.org/10.1126/sciadv.aau4578
Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA (2013) Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2:587. https://doi.org/10.1186/2193-1801-2-587
Shi S, Nuccio EE, Shi ZJ, He Z, Zhou J, Firestone MK (2016) The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol Lett 19:926–936. https://doi.org/10.1111/ele.12630
Shi Y, Li Y, **ang X, Sun R, Yang T, He D, Zhang K, Ni Y, Zhu YG, Adams JM, Chun H (2018) Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 6:1–12. https://doi.org/10.1186/s40168-018-0409-4
Shu D, Guo Y, Zhang B, Zhang C, Van Nostrand JD, Lin Y, Zhou J, Wei G (2021) Rare prokaryotic sub-communities dominate the complexity of ecological networks and soil multinutrient cycling during long-term secondary succession in China’s Loess Plateau. Sci Total Environ 774:145737. https://doi.org/10.1016/j.scitotenv.2021.145737
Sinsabaugh RL, Hill BH, Shah JJF (2009) Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 462:795–798. https://doi.org/10.1038/nature08632
Smith-Ramesh LM, Reynolds HL (2017) The next frontier of plant–soil feedback research: unraveling context dependence across biotic and abiotic gradients. J Veg Sci 28:484–494. https://doi.org/10.1111/jvs.12519
Smith SE, Smith FA, Jakobsen I (2003) Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol 133:16–20. https://doi.org/10.1104/pp.103.024380
Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A (2013) Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. https://doi.org/10.1038/ismej.2013.93
Stegen JC, Lin X, Fredrickson JK, Konopka AE (2015) Estimating and map** ecological processes influencing microbial community assembly. Front Microbiol 6:370. https://doi.org/10.3389/fmicb.2015.00370
Sung J, Lee S, Lee Y, Ha S, Song B, Kim T, Waters BM, Krishnan HB (2015) Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci 241:55–64. https://doi.org/10.1016/j.plantsci.2015.09.027
Tang H, Zhang N, Ni H, Xu X, Wang X, Sui Y, Sun B, Liang Y (2021) Increasing environmental filtering of diazotrophic communities with a decade of latitudinal soil transplantation. Soil Biol Biochem 154:108119. https://doi.org/10.1016/j.soilbio.2020.108119
Tang X, Bernard L, Brauman A, Daufresne T, Deleporte P, Desclaux D, Souche G, Placella SA, Hinsinger P (2014) Increase in microbial biomass and phosphorus availability in the rhizosphere of intercropped cereal and legumes under field conditions. Soil Biol Biochem 75:86–93. https://doi.org/10.1016/j.soilbio.2014.04.001
Thiem D, Gołębiewski M, Hulisz P, Piernik A, Hrynkiewicz K (2018) How does salinity shape bacterial and fungal microbiomes of Alnus glutinosa roots? Front Microbiol 9:651. https://doi.org/10.3389/fmicb.2018.00651
Tian P, Lu H, Feng W, Guan Y, Xue Y (2020) Large decrease in streamflow and sediment load of Qinghai-Tibetan Plateau driven by future climate change: a case study in Lhasa River Basin. Catena 187:104340. https://doi.org/10.1016/j.catena.2019.104340
Tripathi BM, Stegen JC, Kim M, Dong K, Adams JM, Lee YK (2018) Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J 12:1072–1083. https://doi.org/10.1038/s41396-018-0082-4
Upton RN, Bach EM, Hofmockel KS (2019) Spatio-temporal microbial community dynamics within soil aggregates. Soil Biol Biochem 132:58–68. https://doi.org/10.1016/j.soilbio.2019.01.016
Van Breemen N, Finzi AC (1998) Plant-soil interactions: ecological aspects and evolutionary implications. Biogeochemistry 42:1–19. https://doi.org/10.1023/A:1005996009413
Van Der Heijden MG, Bardgett RD, Van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310. https://doi.org/10.1111/j.1461-0248.2007.01139.x
Van der Putten WH, Bardgett RD, Bever JD, Bezemer TM, Casper BB, Fukami T, Kardol P, Klironomos JN, Kulmatiski A, Schweitzer JA, Suding KN, Van de Voorde TFJ, Wardle DA (2013) Plant–soil feedbacks: the past, the present and future challenges. J Ecol 101:265–276. https://doi.org/10.1111/1365-2745.12054
Wagg C, Bender SF, Widmer F, Van Der Heijden MG (2014) Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc Natl Acad Sci U S A 111:5266–5270. https://doi.org/10.1073/pnas.1320054111
Wagg C, Schlaeppi K, Banerjee S, Kuramae EE, van der Heijden MG (2019) Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat Commun 10:4841. https://doi.org/10.1038/s41467-019-12798-y
Wahdan SFM, Heintz-Buschart A, Sansupa C, Tanunchai B, Wu YT, Schädler M, Noll M, Purahong W, Buscot F (2021) Targeting the active rhizosphere microbiome of Trifolium pratense in grassland evidences a stronger-than-expected belowground biodiversity-ecosystem functioning link. Front Microbiol 12:629169. https://doi.org/10.3389/fmicb.2021.629169
Wan W, He D, Li X, **ng Y, Liu S, Ye L, Njoroge DM, Yang Y (2021a) Adaptation of phoD-harboring bacteria to broader environmental gradients at high elevations than at low elevations in the Shennongjia primeval forest. Geoderma 401:115210. https://doi.org/10.1016/j.geoderma.2021.115210
Wan W, Liu S, Li X, **ng Y, Chen W, Huang Q (2021b) Dispersal limitation driving phoD-harboring bacterial community assembly: a potential indicator for ecosystem multifunctionality in long-term fertilized soils. Sci Total Environ 754:141960. https://doi.org/10.1016/j.scitotenv.2020.141960
Wang C, Zhang W, Li X, Wu J (2022a) A global meta-analysis of the impacts of tree plantations on biodiversity. Glob Ecol Biogeogr 31:576–587. https://doi.org/10.1111/geb.13440
Wang M, Wu Y, Zhao J, Liu Y, Chen Z, Tang Z, Tian W, ** Y, Zhang J (2022b) Long-term fertilization lowers the alkaline phosphatase activity by impacting the phoD-harboring bacterial community in rice-winter wheat rotation system. Sci Total Environ 821:153406. https://doi.org/10.1016/j.scitotenv.2022.153406
Wang Q, Chen Z, Zhao J, Ma J, Yu Q, Zou P, Lin H, Ma J (2022c) Fate of heavy metals and bacterial community composition following biogas slurry application in a single rice crop** system. J Soils Sediments 22:968–981. https://doi.org/10.1007/s11368-021-03117-4
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York
Wu H, **ang W, Ouyang S, Forrester DI, Zhou B, Chen L, Ge T, Lei P, Chen L, Zeng Y, Song X, Peñuelas J, Peng C (2019) Linkage between tree species richness and soil microbial diversity improves phosphorus bioavailability. Funct Ecol 33:1549–1560. https://doi.org/10.1111/1365-2435.13355
Wu MH, Chen SY, Chen JW, Xue K, Chen SL, Wang XM, Chen T, Kang SC, Rui JP, Thies JE, Bardgett RD, Wang YF (2021) Reduced microbial stability in the active layer is associated with carbon loss under alpine permafrost degradation. Proc Natl Acad Sci U S A 118:e2025321118. https://doi.org/10.1073/pnas.2025321118
** N, Chu C, Bloor JM (2018) Plant drought resistance is mediated by soil microbial community structure and soil-plant feedbacks in a savanna tree species. Environ Exp Bot 155:695–701. https://doi.org/10.1016/j.envexpbot.2018.08.013
**ong W, Ni P, Chen Y, Gao Y, Shan B, Zhan A (2017) Zooplankton community structure along a pollution gradient at fine geographical scales in river ecosystems: the importance of species sorting over dispersal. Mol Ecol 26:4351–4360. https://doi.org/10.1111/mec.14199
Xue Y, Chen L, Zhao Y, Feng Q, Li C, Wei Y (2022) Shift of soil fungal communities under afforestation in Nanliu River Basin, southwest China. J Environ Manage 302:114130. https://doi.org/10.1016/j.jenvman.2021.114130
Yang Y, Li T, Wang Y, Dou Y, Cheng H, Liu L, An S (2021) Linkage between soil ectoenzyme stoichiometry ratios and microbial diversity following the conversion of cropland into grassland. Agric Ecosyst Environ 314:107418. https://doi.org/10.1016/j.agee.2021.107418
Yu X, Polz MF, Alm EJ (2019) Interactions in self-assembled microbial communities saturate with diversity. ISME J 13:1602–1617. https://doi.org/10.1038/s41396-019-0356-5
Yuan MM, Kakouridis A, Starr E, Nguyen N, Shi S, Pett-Ridge J, Nuccio E, Zhou J, Firestone M (2021) Fungal-bacterial cooccurrence patterns differ between arbuscular mycorrhizal fungi and nonmycorrhizal fungi across soil niches. Mbio 12:e03509-e3520. https://doi.org/10.1128/mBio.03509-20
Zhang H, Wu X, Li G, Qin P (2011) Interactions between arbuscular mycorrhizal fungi and phosphate-solubilizing fungus (Mortierella sp.) and their effects on Kostelelzkya virginica growth and enzyme activities of rhizosphere and bulk soils at different salinities. Biol Fertil Soils 47:543–554. https://doi.org/10.1007/s00374-011-0563-3
Zhang L, Sun P, Huettmann F, Liu S (2022a) Where should China practice forestry in a warming world? Glob Chang Biol 28:2461–2475. https://doi.org/10.1111/gcb.16065
Zhang Z, Lu Y, Wei G, Jiao S (2022b) Rare species-driven diversity-ecosystem multifunctionality relationships are promoted by stochastic community assembly. Mbio 13:e00449-e522. https://doi.org/10.1128/mbio.00449-22
Zhao H, Brearley FQ, Huang L, Tang J, Xu Q, Li X, Huang Y, Zou S, Chen X, Hou W, Pan L, Dong K, Jiang G, Li N (2022) Abundant and rare taxa of planktonic fungal community exhibit distinct assembly patterns along coastal eutrophication gradient. Microb Ecol 1–13. https://doi.org/10.1007/s00248-022-01976-z
Zhou J, Deng Y, Zhang P, Xue K, Liang Y, Van Nostrand JD, Yang Y, He Z, Wu L, Stahl DA, Hazen TC, Tiedje JM, Arkin AP (2014) Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc Natl Acad Sci U S A 111:E836–E845. https://doi.org/10.1073/pnas.1324044111
Zhou J, Ning D (2017) Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-00017. https://doi.org/10.1128/MMBR.00002-17
Zhou Z, Zheng M, **a J, Wang C (2022) Nitrogen addition promotes soil microbial beta diversity and the stochastic assembly. Sci Total Environ 806:150569. https://doi.org/10.1016/j.scitotenv.2021.150569
Zhu F, Lin X, Guan S, Dou S (2022a) Deep incorporation of corn straw benefits soil organic carbon and microbial community composition in a black soil of Northeast China. Soil Use Manag 38:1266–1279. https://doi.org/10.1111/sum.12793
Zhu M, Cao X, Guo Y, Shi S, Wang W, Wang H (2022b) Soil P components and soil fungi community traits in poplar shelterbelts and neighboring farmlands in northeastern China: total alterations and complex associations. Catena 218:106531. https://doi.org/10.1016/j.catena.2022.106531
Acknowledgements
This work was supported by the Second Tibetan Plateau Scientific Expedition and Research Program (2019QZKK0404), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA20020401), Science and Technology Major Project of Tibetan Autonomous Region of China (XZ202201ZD0005G02) and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
R.X.L. performed the experiments, created the figures, and wrote the manuscript. Y.Y., Z.A.G., and Q.L. investigated the data. S.Z. conceived the original research plans, designed the experiments and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Responsible Editor: Enrique Valencia.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, R., Yao, Y., Guo, Z. et al. Plantation rhizosphere soil microbes promote soil‒plant phosphorus feedback on the Tibetan Plateau. Plant Soil (2023). https://doi.org/10.1007/s11104-023-05939-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11104-023-05939-2