Abstract
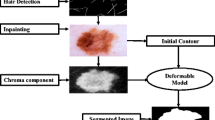
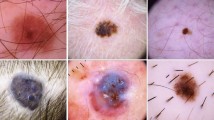
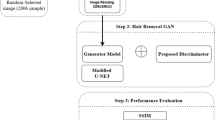
Dermoscopy is commonly used for diagnosing skin cancer in its early stages. However, hair structures in dermoscopy images can negatively affect diagnosis. A proposed algorithm based on conventional image processing methods removes hair structures from these images. Removing hair structures is crucial for constructing accurate computer-aided diagnosis systems for skin cancer. To eliminate hair structures from dermoscopy images, a hair removal algorithm based on conventional image processing methods has been proposed in this study. The proposed algorithm was compared to the existing hair removal algorithms in literature and tested on the ISIC2018 dataset using the AlexNet architecture. The effects on computer-aided systems were assessed by training on images with and without hair. Results show that the algorithm performs well in removing hairs compared to previous studies and improves classification performance. Upon comparison with other literature studies, the recommended algorithm has exhibited consistently high performance, usually ranking among the top two performers in the rank analysis. Additionally, the integration of the suggested algorithm has led to improvements in the performance metrics of the AlexNet architecture, with increases of 0.9% in accuracy, 1.4% in sensitivity, 0.6% in specificity, and 1.06 in F1 score. The performance of the suggested algorithm indicates its potential as a practical and effective tool in clinical settings.








Similar content being viewed by others
Data Availability
The data that support the findings of this study are available in ISIC2018 and Dermaweb, reference number [10], [15] and [40]. These data were derived from the following resources available in the public domain: ISIC2018, https://challenge.isic-archive.com/data/#2018; Dermaweb, http://dermaweb.uib.es/hair-removal-benchmarks/.
References
Abbas Q, Celebi ME, García IF (2011) Hair removal methods: a comparative study for dermoscopy images. Biomed Signal Process Control 6:395–404. https://doi.org/10.1016/j.bspc.2011.01.003
Ariff NAM, Ismail AR (2023) Study of Adam and Adamax optimizers on AlexNet architecture for voice biometric authentication system. Proc 2023 17th Int Conf Ubiquitous Inf Manag Commun IMCOM 2023. https://doi.org/10.1109/IMCOM56909.2023.10035592
Arora G, Dubey AK, Jaffery ZA, Rocha A (2022) A comparative study of fourteen deep learning networks for multi skin lesion classification (MSLC) on unbalanced data. Neural Comput Appl 1–27. https://doi.org/10.1007/S00521-022-06922-1
Attia M, Hossny M, Nahavandi S, Yazdabadi A (2017) Skin melanoma segmentation using recurrent and convolutional neural networks. Proc - Int Symp Biomed Imaging, pp 292–296. https://doi.org/10.1109/ISBI.2017.7950522
Attia M, Hossny M, Zhou H et al (2019) Digital hair segmentation using hybrid convolutional and recurrent neural networks architecture. Comput Methods Programs Biomed 177:17–30. https://doi.org/10.1016/J.CMPB.2019.05.010
Berry K, Butt M, Kirby JS (2017) Influence of information framing on patient decisions to treat actinic keratosis. JAMA Dermatol 153:421–426. https://doi.org/10.1001/JAMADERMATOL.2016.5245
Bibiloni P, González-Hidalgo M, Massanet S (2017) Skin hair removal in dermoscopic images using soft color morphology. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics), pp 322–326. https://doi.org/10.1007/978-3-319-59758-4_37
Brantsch KD, Meisner C, Schönfisch B et al (2008) Analysis of risk factors determining prognosis of cutaneous squamous-cell carcinoma: a prospective study. Lancet Oncol 9:713–720. https://doi.org/10.1016/S1470-2045(08)70178-5
Chadebec C, Thibeau-Sutre E, Burgos N, Allassonniere S (2023) Data augmentation in high dimensional low sample size setting using a geometry-based variational autoencoder. IEEE Trans Pattern Anal Mach Intell 45:2879–2896. https://doi.org/10.1109/TPAMI.2022.3185773
Codella NCF, Gutman D, Celebi ME et al (2018) Skin lesion analysis toward melanoma detection: a challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). Proc - Int Symp Biomed Imaging, pp 168–172. https://doi.org/10.1109/ISBI.2018.8363547
Davidson DW, Fröjdh C, O’Shea V et al (2003) Limitations to flat-field correction methods when using an X-ray spectrum. Nucl Instruments Methods Phys Res Sect A Accel Spectrometers, Detect Assoc Equip 509:146–150. https://doi.org/10.1016/S0168-9002(03)01563-8
Gao J, Pan JH, Zhang SJ, Zheng WS (2023) Automatic modelling for interactive action assessment. Int J Comput Vis 131:659–679. https://doi.org/10.1007/S11263-022-01695-5
Gaulin C, Sebaratnam DF, Fernández-Peñas P (2014) Quality of life in non-melanoma skin cancer. Australas J Dermatol 56:70–76. https://doi.org/10.1111/AJD.12205
Hasan MK, Elahi MTE, Alam MA et al (2022) DermoExpert: skin lesion classification using a hybrid convolutional neural network through segmentation, transfer learning, and augmentation. Informatics Med Unlocked 28:100819. https://doi.org/10.1016/J.IMU.2021.100819
Home | DermaWeb. http://dermaweb.uib.es/. Accessed 13 Jun 2022
Hosny KM, Kassem MA (2022) Refined residual deep convolutional network for skin lesion classification. J Digit Imaging 35:258–280. https://doi.org/10.1007/S10278-021-00552-0
Hosny KM, Kassem MA, Foaud MM (2019) Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS One 14:e0217293. https://doi.org/10.1371/JOURNAL.PONE.0217293
Huang A, Kwan SY, Chang WY et al (2013) A robust hair segmentation and removal approach for clinical images of skin lesions. Proc Annu Int Conf IEEE Eng Med Biol Soc EMBS, pp 3315–3318. https://doi.org/10.1109/EMBC.2013.6610250
Karia PS, Han J, Schmults CD (2013) Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol 68:957–966. https://doi.org/10.1016/J.JAAD.2012.11.037
Kassem MA, Hosny KM, Damaševičius R, Eltoukhy MM (2021) Machine learning and deep learning methods for skin lesion classification and diagnosis: a systematic review. Diagnostics 11:1390. https://doi.org/10.3390/DIAGNOSTICS11081390
Kiani K, Sharafat AR (2011) E-shaver: an improved DullRazor® for digitally removing dark and light-colored hairs in dermoscopic images. Comput Biol Med 41:139–145. https://doi.org/10.1016/J.COMPBIOMED.2011.01.003
Koehoorn J, Sobiecki AC, Boda D et al (2015) Automated digital hair removal by threshold decomposition and morphological analysis. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 9082:15–26. https://doi.org/10.1007/978-3-319-18720-4_2
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60:84–90. https://doi.org/10.1145/3065386
Lee T, Ng V, Gallagher R et al (1997) Dullrazor®: a software approach to hair removal from images. Comput Biol Med 27:533–543. https://doi.org/10.1016/S0010-4825(97)00020-6
Leiter U, Eigentler T, Garbe C (2014) Epidemiology of skin cancer. Adv Exp Med Biol 810:120–140. https://doi.org/10.1007/978-1-4939-0437-2_7
Li W, Joseph Raj AN, Tjahjadi T, Zhuang Z (2021) Digital hair removal by deep learning for skin lesion segmentation. Pattern Recognit 117:107994. https://doi.org/10.1016/J.PATCOG.2021.107994
Mahbod A, Schaefer G, Wang C et al (2020) Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput Methods Programs Biomed 193:105475. https://doi.org/10.1016/j.cmpb.2020.105475
Mat Ariff NA, Ismail AR, Aziz NA, Amir Hussin AA (2022) Analysis of optimizers on AlexNet architecture for face biometric authentication system. 2022 Int Conf Inf Technol Res Innov ICITRI 2022, pp 24–29. https://doi.org/10.1109/ICITRI56423.2022.9970238
Mathews MR, Anzar SM (2021) A comprehensive review on automated systems for severity grading of diabetic retinopathy and macular edema. Int J Imaging Syst Technol 31:2093–2122. https://doi.org/10.1002/IMA.22574
Model MA, Burkhardt JK (2001) A standard for calibration and shading correction of a fluorescence microscope. J Quant Cell Sci 44:309–316. https://doi.org/10.1002/1097-0320(20010701)44:3%3c179::AID-CYTO1110%3e3.0.CO;2-3
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9:62–66. https://doi.org/10.1109/tsmc.1979.4310076
Pereira PMM, Fonseca-Pinto R, Paiva RP et al (2020) Dermoscopic skin lesion image segmentation based on local binary pattern clustering: comparative study. Biomed Signal Process Control 59:101924. https://doi.org/10.1016/j.bspc.2020.101924
Ramella G (2021) Hair removal combining saliency, shape and color. Appl Sci 11:447. https://doi.org/10.3390/APP11010447
Seibert JA, Boone JM, Lindfors KK (1998) Flat-field correction technique for digital detectors. Med Imaging 1998 Phys Med Imaging 3336:348–354. https://doi.org/10.1117/12.317034
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics. CA Cancer J Clin 72:7–33. https://doi.org/10.3322/CAAC.21708
Smith L, MacNeil S (2011) State of the art in non-invasive imaging of cutaneous melanoma. Ski Res Technol 17:257–269. https://doi.org/10.1111/J.1600-0846.2011.00503.X
Süzme NÖ, Güraksın GE (2020) Comparison of image quality measurements in threshold determination of most popular gradient based edge detection algorithms based on particle swarm optimization. Lect Notes Data Eng Commun Technol 43:171–181. https://doi.org/10.1007/978-3-030-36178-5_14
Talavera-Martínez L, Bibiloni P, González-Hidalgo M (2020) An encoder-decoder CNN for hair removal in dermoscopic ımages. Ar**v. https://doi.org/10.48550/arxiv.2010.05013
Toossi MTB, Pourreza HR, Zare H et al (2013) An effective hair removal algorithm for dermoscopy images. Ski Res Technol 19:230–235. https://doi.org/10.1111/SRT.12015
Tschandl P, Rosendahl C, Kittler H (2018) Data descriptor: the HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5:1–9. https://doi.org/10.1038/sdata.2018.161
**e FY, Qin SY, Jiang ZG, Meng RS (2009) PDE-based unsupervised repair of hair-occluded information in dermoscopy images of melanoma. Comput Med Imaging Graph 33:275–282. https://doi.org/10.1016/J.COMPMEDIMAG.2009.01.003
**e F, Li Y, Meng R, Jiang Z (2015) No-reference hair occlusion assessment for dermoscopy images based on distribution feature. Comput Biol Med 59:106–115. https://doi.org/10.1016/J.COMPBIOMED.2015.01.023
Yu Z, Ge Z, Nguyen J et al (2022) Early melanoma diagnosis with sequential dermoscopic images. IEEE Trans Med Imaging 41:633–646. https://doi.org/10.1109/TMI.2021.3120091
Zheng X, Sun H, Lu X, **e W (2022) Rotation-invariant attention network for hyperspectral image classification. IEEE Trans Image Process 31:4251–4265. https://doi.org/10.1109/TIP.2022.3177322
Zhu H, Liu S, Deng L et al (2020) Infrared small target detection via low-rank tensor completion with top-hat regularization. IEEE Trans Geosci Remote Sens 58:1004–1016. https://doi.org/10.1109/TGRS.2019.2942384
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Barın, S., Güraksın, G.E. An improved hair removal algorithm for dermoscopy images. Multimed Tools Appl 83, 8931–8953 (2024). https://doi.org/10.1007/s11042-023-15936-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15936-3