Abstract
Identifying mouth breathing during sleep in a reliable, non-invasive way is challenging and currently not included in sleep studies. However, it has a high clinical relevance in pediatrics, as it can negatively impact the physical and mental health of children. Since mouth breathing is an anomalous condition in the general population with only 2% prevalence in our data set, we are facing an anomaly detection problem. This type of human medical data is commonly approached with deep learning methods. However, applying multiple supervised and unsupervised machine learning methods to this anomaly detection problem showed that classic machine learning methods should also be taken into account. This paper compared deep learning and classic machine learning methods on respiratory data during sleep using a leave-one-out cross validation. This way we observed the uncertainty of the models and their performance across participants with varying signal quality and prevalence of mouth breathing. The main contribution is identifying the model with the highest clinical relevance to facilitate the diagnosis of chronic mouth breathing, which may allow more affected children to receive appropriate treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The rise of machine learning in sleep research is already revolutionizing the diagnosis of sleep disorders (Arnardottir et al 2021) with the automatic classification of sleep stages (Korkalainen et al 2019) and the detection of respiratory events (Huang and Ma 2021). Machine learning as a part of sleep research and clinical practice can reduce the manual effort of physicians and sleep technologists and increase the well-being of the patient through more precise diagnosis and less invasive sleep measurements (Biedebach et al 2023). Acquiring labels for one night of sleep recording requires 2–3 h of manual review by a sleep technologist who is an expert in the field, which is both time-consuming and expensive (Arnardottir et al 2022). Some potentially important events during sleep, such as mouth breathing, are often not labeled at all. Mouth breathing is a particular problem for children, since they may face developmental issues if they breath through their mouth during sleep, at an early age (Gozal 1998). Children with sleep-disordered breathing can suffer from serious long-term implications if their condition is not recognized and appropriately treated (Marcus 2001). In fact, chronic mouth breathing can lead to obstructive sleep apnea (Izu et al 2010), learning disorders, (Fensterseifer et al 2013) and a malformation of the child’s jaw area (Denotti et al 2014). The task of identifying mouth breathing with machine learning is challenging, since we are facing an anomaly detection problem. Healthy children usually breathe through their nose, which makes mouth breathing an anomalous behavior (Lee et al 2015). In this paper, we analyze mouth breathing of children, in a highly imbalanced data set, which included only few mouth breathing sequences because most of the children did not breathe through the mouth at all. From the 20 labeled recordings, the child with the highest duration of mouth breathing had a total length of 1980 s or 33 min of mouth breathing in a night with 10 h of sleep. Overall, the data set that has only 2.4% positive examples. Therefore, we assume that mouth breathing is an anomaly. In this paper, we aimed to reduce the manual effort for identifying mouth breathing in order to enable a more efficient diagnosis of the condition, which will hopefully enable more children to receive treatment before these health implications surface. There are two central challenges to identifying mouth breathing. Firstly, even for the sleep technologist, it is difficult to make a clear distinction between mouth and nose breathing, since the boundary is blurred. In general, there is no mouth breathing, if the mouth is closed and the air is solely flowing through the nose. However, the same generalization is not valid for the other way around; air can flow both through the nose and the mouth when there is mouth breathing (Koutsourelakis et al 2006). Secondly, it is challenging to acquire a sufficient amount of labeled mouth breathing events, since mouth breathing is an anomalous behavior in the healthy population and they are usually not labeled. As a result, the labeled recording might include only a few or no mouth breathing events at all.
When approaching this type of human medical data with machine learning, different rules than for tabular data apply. We need to consider that each training example is a breathing sequence that belongs to a certain unique individual. This impacts both the training and testing of the machine learning model. Splitting the data with a common random train test split, could lead to a data leakage problem during the model training. Peralta et al (2021) show in a systematic review in machine learning for deep brain stimulation, that more than half of the papers in this field do not do a patient-wise validation. Therefore, we created the train, test and validation set by separating the data by participants as shown by Oner et al (2020). This is in line with the practical aspects of implementing the machine learning model in clinical practice, where the data of a new patient is fully separated from the data the model was trained on. The same logic holds for the evaluation of a machine learning model with human medical data. Evaluating the performance of the model on participants separately can reveal whether the model performance varies among different groups of participants with certain characteristics and whether the model can generalize on all patients.
In this paper, we aimed to find the best way to predict pediatric mouth breathing during sleep by comparing different supervised and unsupervised machine learning models. The data set included sleep recordings of 111 participants using oronasal cannulas. We transformed the data set, by first splitting the full sleep recordings into 10-second subsequences. We trained the model on multiple signals of the sleep recordings including thorax and abdomen movement, oral pressure, nasal pressure, blood oxygen saturation, heart rate, audio volume and position. We tested the model using a leave-one-out cross validation and chose the model with the highest clinical relevance. The main contributions of this paper are three-fold: (i) to illustrate the challenges and required preprocessing steps for applying machine learning to sleep data, (ii) to identify the models with highest clinical value, and (iii) to contribute to the discourse of when deep learning is needed and when simplicity is key.
Our work makes an important contribution to the field of sleep research, as we show that mouth breathing during sleep can be automatically identified with machine learning, which allows a faster diagnosis of mouth breathing. Most significantly, our work contributes to machine learning as we show the challenges of working with human medical data and outline a sensible preprocessing, training and evaluation method to counteract them. Importantly, we approached this problem with different machine learning methods, including a naive baseline, a classic machine learning model, time series classifiers, deep learning models and an unsupervised anomaly detection method. Evaluating these different methods in a leave-one-out cross validation showed their performance across the whole population and on an individual basis, which raised the question whether classic methods are preferable over deep learning for anomaly detection in sleep. The rest of this paper is organized as follows.
In the next section, we summarize the existing literature related to unsupervised anomaly detection and mouth breathing identification. Then, we describe our proposed methodology for automatic detection of mouth breathing events, followed by a presentation of our results. The paper ends with a discussion of the implications of our contribution and steps for future work.
2 Related work
2.1 Time series anomaly detection
Time series anomaly detection, as a subfield of anomaly detection, has been studied in literature. Common application fields of anomaly detection are healthcare (Chauhan and Vig 2015), financial fraud (Fu et al 2016), robotics (Park et al 2018) or network intrusion (Leung and Leckie 2005). Literature reviews (Blázquez-García et al 2021; Chalapathy and Chawla 2019) show the broad range of methods and application fields of time series anomaly detection. Experimental comparisons have been conducted and compare the performance of both supervised (Freeman et al 2021) and unsupervised (Rewicki et al 2023) anomaly detection methods on time series bench marking data sets. This paper did not aim to do a exhaustive experimental comparison as the aforementioned papers, but instead to provide a detailed understanding of the application of anomaly detection on sleep data and show the challenges of detecting anomalies in this type of human bio signals.
2.2 Identifying mouth breathing
The literature on automatic identification of mouth breathing during sleep is scarce. In the existing literature, mouth breathing is typically identified with questionnaires (Sano et al 2018) and direct observation (de Castilho et al 2016). Mouth breathing measurement is still not commonly included in a standard polysomnography recording and moreover, not manually labelled as a standard practice. One reason is the lack of a reliable and non-invasive measurement device. An oronasal cannula can separate the airflow, but is easily misplaced or fully removed during sleep. An orosonasal thermistor captures the oral flow by measuring the temperature above the mouth (Koutsourelakis et al 2006), but thermistors have a low signal quality (Sabil et al 2019). A specialized mask can be used to separate the breathing channels, but wearing the device may bias the breathing and it is not suitable for children (Hudgel et al 1984). Curran et al. differentiated between the breathing channels by processing the sound during sleep (Curran et al 2012). They applied a Fast Fourier transformation to the raw audio signal, split it into windows of of 5–15 s and trained a deep neural network on this input. Their work is only comparable to a certain extend, because their data stemmed from recordings during awake in a controlled environment where the participants were instructed to breath through their mouth or their nose with an airflow of 1.7 l per s. The data used in our research reflects the real-world conditions of noisy signals and uncontrolled airflow during sleep.
3 Method
3.1 Data
This paper is based on a a comprehensive data setFootnote 1 that includes paediatric sleep recordings, including 10–13-year-old children with parent reported sleep-disordered breathing symptoms and a gender and age-matched control group. The study cohort consists of 111 children from the Icelandic EuroPrevall-iFAAM birth cohort research (Grabenhenrich et al 2020; Keil et al 2010; Sigurdardóttir et al 2021) conducted at the Landspitali University Hospital. The sleep recording was done with a Nox Medical A1 polysomnography (PSG) device.
PSG is the continuous recording of physiologic activity during sleep. The measurement includes the following sensors: Electroencephalogram (EEG), electrooculography (EOG), chin and leg electromyography (EMG), electrocardiography (ECG), pulse oximeter, microphone, electrodermal activity (EDA) sensor, and accelerometry measuring the movement and body position. Thoracic and abdominal respiratory inductance plethysmography (RIP) belts measure the inflation and deflation of the chest during breathing and an oronasal cannula with separate pressure outputs, monitors the nasal and oral airflow, respectively (Markun and Sampat 2020). A visualization of a PSG set up with an oronasal cannula can be seen in Fig. 1. The PureFlow oronasal cannula by Braebon is specially designed to capture the oral flow and nasal flow separately and was utilized in this study, but is not included in a standard PSG. The PSG was set up at the hospital by sleep technologists, but the participants slept at home and returned the devices the next morning (Kainulainen et al 2021).
Each PSG recording was approximately 8 h long, containing 84 different signals in total. We focused on the nasal and oral flow as well as the thorax and abdomen movement, which measured breathing or movement. Additionally, we included blood oxygen saturation, the audio volume, the heart rate, and body position in the analysis. Two exemplary sequences of the respiratory signals can be seen in Fig. 2, where the thorax movement is colored in light blue, the abdomen movement dark blue, the nasal flow dark green, and the oral flow light green. The top shows a typical nose breathing sequence and the bottom shows a typical mouth breathing sequence. In the mouth breathing sequence, the oral flow shows higher amplitudes than the nasal flow and the amplitudes of the nasal flow are lower than during nose breathing. This behavior is typical for mouth breathing, but cannot be generalized to all mouth breathing sequences. There were 111 recordings, of which 20 have been manually labeled by a sleep technologist. The manual labels were based on the oral and nasal flow signals. 10 recordings were manually chosen to have 5 healthy children and 5 children with sleep disordered breathing. The latter 10 recordings were chosen because the parent-reported information from the questionnaires indicated mouth breathing.
3.2 Preprocessing
The data of PSG recordings were saved in the.edf standard format, an open-source file format commonly used for medical data in Europe. It is designed for multi-channel medical time series and allows different sampling frequencies for each signal (Kemp et al 1992). For each PSG, we extracted the signals of interest and each signal’s sampling frequency. Some PSGs had faulty or missing recordings from the RIP belts or the cannula, therefore each recording was visually checked for completeness. During this process, two labeled studies were removed due to low signal quality or measurement errors, which led to a total of 18 eligible labeled sleep studies for this paper.
The four respiratory signals have a sampling frequency of 200 Hz. As the average duration of one study is 8 h, one PSG contains on average 5,760,000 values per signal. Therefore, the full data set quickly becomes complex. To process this large amount of data, we faced a trade-off between run-time and the completeness of the data representation. We downsampled the signals to a sampling frequency of 10 Hz, because this reduced the complexity, but still captured the relevant features in the data. We furthermore upsampled the oxygen saturation and heart rate with a sampling frequency of 3 Hz to have a common sample rate (Gao et al 2018). The differences between too simplified downsampling to 1 Hz, the chosen downsampling to 10 Hz, and the original sampling frequency of 200 Hz can be seen in Fig. 3.
The signals have different scales, as the oral flow has a range from approximately −2 cm H2O to 1 cm H2O, while the thorax and abdomen only range between \(-\)0.0005 V and 0.0005 V. As some models are sensitive to different sized scales, we prevented the signals with larger scales to out rule the signals with smaller scales, by scaling the data to the same range using the scikit-learn StandardScaler. In order to treat this data set as a time series classification problem, we split the data into sequences of 10 s. One respiration cycle, i.e., inhalation and exhalation, of children that are 6 years and older usually has a duration of 2–5 s during sleep (Fleming et al 2011). Choosing an interval of 10 s guarantees that the interval contains at least one full breath and up to 5 full breaths. We did not choose a longer duration than 10 s, to do the classification as granular as possible and keep the complexity of the data, i.e., the length of the time series, as low as possible. We tested both disjoint splits and sliding window splits but as no visible difference in model performance was perceived, we chose the less complex method of disjoint splits. The target variable ’breathing channel’ was labeled as 1 for mouth breathing or 0 for nose breathing. It was assigned to each sequence based on the annotation file. To be considered as target class 1, the sequence had to contain at least 3 s (the average length of one respiration cycle) of mouth breathing according to the manual labels by the sleep technologist.
3.3 Model training and hyperparameter optimization
The recording of each participant included approximately 2500 sequences. Since each participant has individual characteristics, their sleep recordings have individual characteristics as well, which is why we used a leave-one-out cross validation for model training and evaluation, i.e., we trained the model on all participants but one and tested the model performance on the test individual. This way, we ensured that no data of the test individual leaked into the model training. For the hyperparameter optimization, a validation set of 3 participants was separated from the rest of the participants. These recordings were only used for hyperparameter optimization and were not included in the leave-one-out cross validation. The validation participants included one with more than 10 mouth breathing sequences, one with zero mouth breathing sequences and one with low signal quality to represent different types of recordings that were present in the data set. We optimized the hyperparameters of all deep learning models on this separate validation set with a keras RandomSearch. For this we defined a grid of possible values for the number of filters, the kernel size, the dropout rate and the learning rate, from which the RandomSearch randomly selected parameter combinations. The hyperparameter optimization has been conducted in the same method and same extend for all deep learning models. The unsupervised deep learning models were optimized to achieve a maximal accuracy in the validation set. The autoencoder was optimized with a a custom loss function to to maximize the average reconstruction error of mouth breathing divided by the average reconstruction error or nose breathing. The hyperparameters of the feature-based model were optimized with a RandomSearch as well. Here, we tuned the learning rate and number of estimators. The hyperparameters for the time series classifiers were set through testing different values on this validation set manually.
3.4 Machine learning methods
As a comparison of multiple machine learning methods, we compare three different time series classifiers and two supervised deep learning models using the raw time series as an input. Ultimately, we propose a reconstruction-based method and feature-based method. Each of the methods will be described in the following.
3.4.1 Supervised time series and deep learning models
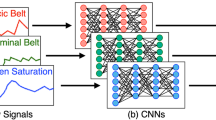
All three time series models work with different representations of the data. This includes using the full sequence, fixed intervals, or dynamic shapelets in the training (Bagnall et al 2017). A brief explanation of each model and the selected parameters can be seen in Table 1. We also test two deep learning models, since they can handle multivariate time series, which allows them to capture the interaction between signals.
3.4.2 Reconstruction-based anomaly detection
Autoencoders are commonly implemented with multi-layer neural networks. They learn an encoding and a decoding function using an iterative optimization process. The data is passed through the network, the reconstruction error is calculated and at each iteration, the weights of the network are updated (LeCun et al 2015). In a convolutional autoencoder, convolutional layers are included in the encoder and deconvolutional layers in the decoder of the neural network (Ribeiro et al 2018). Convolutional layers transform the data by sliding a filter over the time series. Several filters of different sizes can be applied to learn multiple discriminative features from the input time series. The deconvolutional layers, or transposed convolutions, work by the same principle but swap the forward and backward passes of the convolution. Average- or MaxPooling reduces the length of a time series by aggregating it with a sliding window (Fawaz et al 2019). The hidden layers aim to separate relevant and irrelevant features, which can hide the presence of anomalies (Chalapathy and Chawla 2019). In the encoder, lowering the dimensionality of the input with the convolutional layers, creates a bottleneck after which ideally only the most explanatory parts of the data remain. In the decoder, the transposed convolutions increase the dimensionality of the data back into its original shape. The new representation, i.e the reconstructed input, will naturally differ from the original representation. However, this deviation is encouraged since it did not happen at random, but is a result of the learned weights of the autoencoder. Autoencoders are trained with the objective of minimizing the reconstruction error, i.e., the error between the original input and the reconstructed output (Li et al 2020). As the majority of the examples in the training data belong to the normal class, the autoencoder will mainly learn the properties of this normal class (Chalapathy and Chawla 2019). Reconstruction-based anomaly detection relies on the assumption that the reconstruction of anomalies is less accurate than the reconstruction of normal instances. As a result, the reconstruction error is higher for anomalous examples, which allows us to use it as an anomaly score and detect anomalies in a fully unsupervised way (Chandola et al 2009).
We used the Root Mean Squared Error (RMSE) as distance metric for defining the reconstruction error, because the input and output of our model were multivariate time series. We chose RMSE above other distance metrics as it gives relatively high weight to large errors. For each 10-second interval, we calculate the RSME by averaging the squared root of the distance between the original and reconstructed signal at all 100 time steps. Equation 1 shows how the RMSE for signal j is calculated over all n=100 time steps:
In the model training, we used the average RMSE of all included signals as the loss function for optimization. In the reconstruction-based anomaly detection, we used the RMSEs of the individual signals as new features. In order to use these new features for reconstruction-based anomaly detection, we need to define a classification threshold t. All examples with a higher reconstruction error than t are classified as anomalies and all examples with a lower reconstruction error than t were classified as normal instances. Hence, we classify all examples above t as mouth breathing, and all examples below t as nose breathing. Defining an appropriate threshold is crucial for the success of the anomaly detection. We ran experiments with different approaches of setting the threshold to find the one which achieves the most accurate classification. The most straightforward approach is taking the average reconstruction error of all signals. We also took the distribution of the data into account by adding the standard deviation of the reconstruction error to the threshold. Thus, we defined the threshold t as the average reconstruction error plus the average standard deviation over s signals as shown in Eq. 2.
Knowing the reconstruction error of each signal individually, allows us to use subsets of the signals for defining a threshold as well. Figure 4, a correlation matrix between the reconstruction errors and the breathing channel, here referred to as target variable y, shows that some reconstruction errors correlate more with the breathing channel than others. The highest correlation can be seen between y and the oral pressure. For this reason, we propose a second approach of setting the threshold, using only the most discriminative feature, the oral flow, instead of the average. The definition of this threshold t is shown in Eq. 3.
During this experimenting, we observe that sequences with bad signal quality have extremely high overall average reconstruction errors. Hence, we define a threshold that uses both the reconstruction error of the oral flow and the average reconstruction error. We extend the definition from Eq. 3 by adding a constraint on the average reconstruction. Now, we furthermore discard sequences which have a higher average reconstruction error than 99% of all other sequences.
The autoencoder in this paper was implemented with a convolutional neural network using TensorFlow and is built as a keras sequential model. We constructed the autoencoder by starting with a simple set-up, the input layer, one convolutional layer, and one transposed deconvolutional layer followed by a max-pooling- or respectively upsampling layer with all default values. To avoid the risk of overfitting, we added dropout layers for regularization. Then the complexity of the model was gradually increased and an autoencoder with two hidden layers in the encoder and decoder was chosen. One important property of the model is the dimensionality of the latent space. The representation of the data in this state is crucial for the reconstruction and therefore the success of the anomaly detection. A too big latent space prevents the data from learning a model at all. In the most extreme case, with a latent space of the same size as the input space, the reconstruction error is zero and no classification is possible. Choosing a too small latent space is also not recommended, as too much information is lost in the bottleneck. We choose a range of possible filter and kernel sizes that do not allow a too small or too big latent space in the hyperparameter optimization. The full autoencoder architecture can be found in Fig. 9 in the appendix. The hyperparameter tuning results in an optimal learning rate of 0.001, using the RMSProp optimizer. Finally, we train the model in 50 epochs with a batch size of 256.
We trained the unsupervised model on a bigger train set than the other models, as it additionally included unlabeled recordings, but we tested it on the same test set as the other models. Since the participants of the study were not randomly selected, but consisted of 50% of children with a history of sleep-disordered breathing and 50% of a control group with a history of normal breathing, we did a pre-selection for the train set. This step aimed to lower the number of mouth breathing sequences in the train set to a level that reflects the average population better. This pre-selection was done based on a parental questionnaire regarding the child’s breathing behavior during sleep. If the parents answered that they observed their children slee** with an open mouth, waking up with a dry mouth, or breathing through their mouth during the day, we disqualified the child from the train set. This led to disqualifying 54 children from the training, which approximately reflects the percentage of the study population with abnormal breathing behavior. Including these children in the train set could contradict the assumption that mouth breathing is the rare exception. It is still possible, that mouth breathing was included in these recordings, but we can assume that the proportion of mouth breathing in this subset of recordings was low.
Reconstruction-based anomaly detection can also be implemented as a semi-supervised model. Similar to the unsupervised approach, the autoencoder is trained without using any labels and no labeled anomalous examples are needed. Instead, we use only examples of the normal class for the model training as described in Chalapathy and Chawla (2019). The idea behind a semi-supervised approach is to train the autoencoder only on sequences that certainly do not contain any mouth breathing. This way, we do not have to rely on the assumption that the imbalance in the data set is high enough to disregard the mouth breathing sequences in the training data.
3.4.3 Feature-based classification
As a comparison to the unsupervised deep learning model, we train a classifier which works with simple statistical features. We transform the 3-dimensional time series data set into a 2-dimensional data set with time-independent features. This is done by calculating summary values for each sequence of 100 time steps, including the mean, standard deviation, minimum and maximum of each signal. Additionally, we create two more features based on the oronasal cannula. We calculate the difference between the mean of the oral flow and nasal flow, as well as the difference between the standard deviation of the oral flow and nasal flow, which can be seen in Eq. 4, where n is the number of time steps.
We then applied a feature selection based on the Pearson correlation coefficient between the feature and the target variable. We selected the 10 most correlated features for the model training. Figure 5 shows the correlations of the 10 selected features.
These features were then used as an input for the supervised classification method gradient boosting machine (GBM). A GBM is an ensemble method that commonly excels over other machine learning methods in bench marking studies or practical applications (Natekin and Knoll 2013). Ensemble methods use the combined classification power of multiple individual machine learning classifiers. While other ensemble methods simply average the predictions of multiple classifiers, the strength of a GBM lies in the sequential training of models, which allows it to take the classification errors of the previous models into account. We trained the GBM with a learning rate of 1 and use 1000 estimators.
3.5 Evaluation
We evaluated the models by comparing the predicted labels by our models to the manual labels provided by the sleep technologist. To achieve a comprehensive evaluation of how the models can predict mouth breathing in unseen sleep recordings, we performed a leave-one-out cross validation. This form of cross validation evaluates the models for each sleep recording individually by using the recordings of all children but one for training and the remaining one for testing. This way, we could observe the inter-subject variability and use a higher amount of training data. We calculated the average precision and recall scores as proposed by Forman and Scholz (2010) to avoid bias from the class imbalance in different folds. As we are facing a highly imbalanced classification problem, it does not make sense to consider accuracy as an evaluation measure. Instead, we rely on metrics, which evaluate the classification of the minority class, such as precision, recall, and F1 score. For the final results, we added up the confusion matrices of all folds for each model and calculated the precision, recall and F1 score from the total number of true positives, false positives and false negatives. The individual classification results of the models on each child’s recording can be found in Table 4 in the appendix.
We furthermore calculated the standard deviation of these different metrics across the participants. This showed how much the classification performance varies across participants and therefore how good the model can generalize on test sets with unique characteristics. Another perspective towards the anomaly detection is added when we divide the participants in the test set by a high or low number of mouth breathing, which represents the group of healthy and sleep disordered participants. This analysis reveals which models can handle test sets with close to zero positive examples. In this group we additionally calculate the False Positive Rate (FPR) by dividing the number of false positives by the number of false positives and true negatives. This test shows how strongly a model would overestimate the degree of mouth breathing in an healthy individual.
4 Results
4.1 Naive baseline
As a naive baseline, we do stratified random guessing, which takes the distribution of nose breathing and mouth breathing examples in the train set into account. Each sample in the test set gets the label nose breathing or mouth breathing with a probability that reflects the class distribution. This approach resulted in an F1 score lower than 0.01, which gives a hint at the difficulty of classification in a highly imbalanced data set.
4.2 Overall evaluation
All supervised models were evaluated within the same leave-one-out cross validation as the reconstruction-based anomaly detection to achieve comparability between all models. Table 2 shows the performance of all models evaluated on 15 different test folds using the leave-one-out cross validation. Evaluating the overall performance of the machine learning models, showed that the GBM using statistical features as an input is the best classifier with an F1 score of 0.54. The reconstruction-based classifier has a similar F1 score of 0.508. Comparing the classification accuracy of all supervised models showed, that the deep learning models were not necessarily better than the time series classifiers. The classifiers KNN-DTW and MRSEQL, which can handle multivariate time series as an input, performed better than the TSF, which combines the predictions of the individual time series in an ensemble. The best performing supervised model was MRSEQL, which even exceeded the performance of the deep learning models. Both deep learning models have a lower classification accuracy than the classic feature-based classifier. The recall, i.e., how many of the true mouth breathing sequences were identified as such, and the precision, i.e., how many of the predicted mouth breathing were correct, give an enhanced insight on the model performance. Most machine learning models in the evaluation had a high recall but a low precision. This means they identified many true positives, but also predicted many false positives. The only exceptions to this trend were the deep learning models. Both the RNN and the CNN had a higher precision than recall. Especially the CNN led to a low recall score, as it fails to identify most of the true mouth breathing sequences. Looking at the individual training folds showed, that for some participants the CNN was not able to make any prediction and resulted in an F1 score of 0.
4.3 Individual-level evaluation
Evaluating the performance of the machine learning models on an individual level showed that the classification accuracy of all models varies strongly. The best performing models, the feature-based classifier and the reconstruction-based classifier both had a standard deviation of the F1 score of 0.3. This shows that the models work well for some participants and perform poorly on other participants. Reviewing the participants one by one showed that the performance of all models was weaker for the participants with a lower amount of mouth breathing. This is plausible, as an increased imbalance ratio affects the classifier performance as shown by Lemnaru and Potolea (2011). Furthermore, having evaluation folds with zero positive examples in the test set can naturally only lead to a decrease of precision, recall and F1 score as achieving true positive classifications is impossible in this setting. However, it is a relevant results since these participants with zero or few mouth breathing sequences represent healthy, non-mouth breathing participants, which is the majority of the population. Dividing the test set into participants with more than 10 mouth breathing sequences and lower or equal to 10 mouth breathing sequences shows how each model would perform when classifying healthy or sleep disordered participants.
Figure 6 shows the F1 score of the individual participants as a distribution for each machine learning model. The plot on the left side shows the low or non-mouth breathing participants and the plot on the right side shows the participants with more than 10 mouth breathing sequences. We can see that the CNN, which did not perform well in the overall evaluation with an F1 score of 0.179 was the best performing model for the participants with a low-amount of mouth breathing. The reconstruction-based model and the feature-based model both had a low performance on the low-mouth breathing sequences. However, the autoencoders and the GBM were leading the performance in the high-mouth breathing participants. Both the CNN and the RNN have a False Positive Rate (FPR) of 0.007. This equals to approximately 15 false positives on average in each participant. Even though the feature-based classifier in comparison has a FPR of 0.015 with 33 false positives on average.
4.4 Reconstruction-based anomaly detection
Applying the autoencoder on an unseen test set resulted in a reconstruction error that was indeed higher for mouth breathing than for nose breathing. The average reconstruction error of the anomalous class was twice as high as the average reconstruction error of the normal class. This can be seen in Fig. 7, which shows the distribution of the reconstruction errors of mouth breathing and nose breathing separately.
We can see that most of the nose breathing examples (92.8%) have a reconstruction error below 1. The mouth breathing examples have higher reconstruction errors in a range between 0.5 and 5. This shows different distributions of the reconstruction error of the two classes, which is why we can use the reconstruction error as an anomaly score. However, as the classes do overlap (shown in red in Fig. 7), a perfect separation of normal and anomalous data by threshold was impossible based on the reconstruction error.
Figure 8 shows how precision, recall and F1 score changed by gradually increasing the threshold value. Testing the classification accuracy of the different threshold approaches showed that the way the threshold was defined had a high impact on the classification performance. The estimated thresholds used for classification did not necessarily match the optimal thresholds like the one shown in Fig. 8 as the grey dotted line. Figure 8 shows that even small deviations from this optimal separation led to a strong decrease in classification performance. Hence, the following results show the classification ability of this particular unsupervised classification approach, but do not necessarily reflect the full potential of the autoencoder for reconstruction-based anomaly detection.
The summarized results of evaluating the unsupervised- and semi-supervised autoencoder are shown in Table 3. The classification with the average threshold led to low results even though theoretically a separation of the classes is given as seen in Fig. 7. The average threshold is not only higher for mouth breathing but also in bad signal quality. For this reason, it was not suitable for the unsupervised detection of mouth breathing. The results strongly improved when only the oral threshold is used for the classification. This approach achieved the highest recall of 0.827 in the semi-supervised training. We could further improve the F1 score of this approach by combining the information from the average and mouth breathing reconstruction error. This approach achieved the best overall results in the semi-supervised training with a precision of 0.401, a recall of 0.695 and an F1 score of 0.508.
4.5 Error analysis
In order to gain a deeper understanding of the classification performance, we review a subset of the misclassified sequences of the reconstruction based anomaly detection with a sleep technologist. In particular, we review the false positives, i.e the nose breathing sequences the model labels as mouth breathing. We take the time stamps of a subsample of the test set and review these sequences in the sleep analysis software Noxturnal by Noxmedical. The following reasons for misclassifications were identified:
-
Slight or short mouth breathing: Some sequences show mouth breathing in the manual review but were not labeled as such, because it was only slight mouth breathing. In many of these cases, it was shortly before or after a labeled sequence of mouth breathing. Others were correctly labeled but were disregarded in the preprocessing because the mouth breathing was very short and only sequences with at least three seconds of mouth breathing were considered as mouth breathing sequences.
-
Mouth breathing during awake state: There are multiple sequences that actually had mouth breathing but were not labeled as such by the sleep technologist, because they occurred during an awakening, which is not considered as clinically relevant.
-
Bad signal quality: In some sequences, the sleep technologist cannot decide whether mouth breathing is present, because the signals are noisy or include artifacts. In one sequence it is clearly visible that the oximeter lost contact. In other cases, we assume that the oronasal cannula has moved.
This review shows us that the we cannot always rely on the manual labels as the ground truth. Setting clear borders where mouth breathing starts and stops is a challenge for the human reviewer as well, especially in recordings with bad signal quality. We should keep in mind that assigning the labels is a subjective task and that interrater variability is an ongoing research area in sleep (Danker-hopfe et al 2009). It also reveals a weakness of the preprocessing, which indicates we should lower the required minimum amount of mouth breathing per sequence in future work. Whether we should exclude mouth breathing during awakenings from the evaluation or include information about the sleep stages in the input data remains an open question. Overall, the error analysis also showed that many of the false positives have not been entirely false after all.
5 Discussion
The results demonstrated that machine learning can be used to automatically differentiate between mouth breathing and nose breathing. The comparison of time series classifiers, deep learning models, unsupervised models and a feature-based classifier showed that overall the feature-based classifier was the best performing machine learning method. Evaluating the performance of these models in the leave-one-out cross validation showed that the model performance varied strongly across participants. The two best performing models, the reconstruction-based anomaly detection and the feature-based classifier showed a similar standard deviation. They also showed a similar performance drop on the test set with a low number of mouth breathing in comparison to the test set with participants with a high number of mouth breathing. This showed that both models may overestimate the severity of mouth breathing when used in clinical practice.
To assess whether this classification accuracy is precise enough to replace manual annotation work, we should consider the implications of false positive and false negative classifications for the sleep technologists, as well as the consequences that arise for the child. Classifying too many nose breathing sequences as mouth breathing sequences gives the impression that a child suffers from a condition they do not have or only mildly suffer from. On the contrary, capturing none or too few of the true mouth breathing sequences may lead to underestimating the severity of mouth breathing and preventing the child from receiving the appropriate diagnosis and treatment. Our best performing model has a precision of 44.5% and can identify 70.4% of all mouth breathing sequences. Therefore, it is likely to identify a high percentage of the mouth breathing sequences but may overestimate the mouth breathing. Both the feature-based and reconstruction-based methods have a low precision but high recall. Therefore they could be suitable to support the sleep technologist by highlighting the sequences which are likely to be mouth breathing and leave the final decision to the expert. This approach of supporting the medical staff instead of fully replacing medical staff has shown success when integrating machine learning applications in clinical practice (Henry et al 2022). Whether sleep technologists rely completely on the prediction in the future or use it as a reference value for faster manual review depends on the desired accuracy of the mouth breathing labels, but either way decreases the manual labeling effort.
Applying the reconstruction-based anomaly detection approach on sleep data and observing separation of the classes by reconstruction error shows that this approach is applicable to sleep data. We can see that the unsupervised approach has a lower classification accuracy than the semi-supervised approach. There are several reasons which may account for this gap. Firstly, the remaining mouth breathing sequences in the train set of the unsupervised approach negatively impact the reconstruction-based anomaly detection. This would show that our proposed model strongly relies on the assumption of an imbalanced data set. Secondly, we also include non-labeled recordings in the train set of the unsupervised model. As these have not been reviewed manually, we have no information of the amount of mouth breathing or the signal quality in these recordings. Another limitation of the reconstruction-based anomaly detection is that the autoencoder is not able to differentiate between different types of anomalies. Even though we assume that mouth breathing sequences are anomalies, we cannot assume that all anomalies are mouth breathing. Consequently, the false positives, that are incorrectly classified as mouth breathing partly also point towards other anomalies such as measurement errors, which makes our model less applicable for low quality recordings.
However, the comparison of different methods showed that a classic machine learning approach outperforms the deep learning models. The feature-based classifier simplifies the time series into summary features. This shows that the shape of the signals is not relevant for the classification, but rather their altitude and range. Therefore, our research shows, that for this specific application, deep learning models are not superior to classic machine learning models. This goes in line with previous publications questioning the need for deep learning in other domains (Gunnarsson et al 2021; Shwartz-Ziv and Armon 2022). It is an ongoing debate when and how deep learning is needed. The superior performance of our model in comparison to the model by Curran et al (2012) may arise from including more features than only the audio signal. However, including more signals has not only advantages, as a PSG study is more of an effort than a microphone study. The overall results show, that the signals we included in the model are suitable for identifying mouth breathing. It is surprising that the statistical features and the reconstruction error show different correlation with the target variable mouth breathing, as shown in Figs. 4 and 5. While the reconstruction error of the autoencoder mainly shows correlation of the oral flow and the target variable, the statistical features also shows correlations to the audio volume and the oxygen saturation. One reason for that could be that these signals have different properties and are the blood oxygen and audio volume are more meaningful as summary statistics and the oral flow signal is more meaningful as a raw signal. A mixed approach of inputting the oral flow as a raw signal and the audio and blood oxygen saturation as statistical features could be an interesting approach to pursue in future work.
We need to keep in mind, that machine learning models can identify complex patterns from the training data but do not have human reasoning. In one sleep recording, the oronasal cannula is misplaced in a way that the pressure transducer, which captures the mouth breathing, was placed above the nose. For 6 h, the nose breathing signal is gone, but the mouth breathing is unusually high. Listening to the audio and looking at the unusual patterns of the mouth breathing signal, let the sleep technologist conclude that it was a measurement error, even though it looked like extreme mouth breathing. This is a line of thought that comes naturally to the sleep technologist, but can not be achieved by a machine learning model. For this reason, the ability of our machine learning models is limited by the quality of the sleep recording and can be negatively impacted by measurement errors.
6 Conclusion
These findings are relevant for research focusing on sleep-disordered breathing, because they show that mouth breathing can be automatically identified. Using the signals from the oronasal cannula, thorax and abdomen belts, pulse oximeter, and microphone, our proposed approach can classify mouth breathing with an F1 score of 0.546. This means, that the manual annotation work can be decreased with the use of machine learning. Comparing classic and deep machine learning models, showed that classic methods outperform deep learning and have a higher clinical relevance in this application. The results from the reconstruction-based method showed that we are not dependent on labeled mouth breathing sequences in the training to identify mouth breathing. The results of all machine learning models varied strongly across participants, which highlights the importance of patient-wise evaluation. In future research, we want to test whether these models also work on the data of adults with sleep-disordered breathing. To improve the model performance, the superordinate time series should be taken into account, as a sequence is more likely to be mouth breathing if the preceding and succeeding sequences are mouth breathing as well. The model could be further improved by classifying individual breaths instead of fixed 10-second intervals as proposed in Holm et al (2022).
Notes
The access to this data was granted by the National Bioethics Committee of Iceland. The study was approved by the Data Protection Agency of Iceland and includes written consent from each child’s legal guardian. We cannot make the data publicly available, as is it protected by the ethical approval.
References
Arnardottir ES, Islind AS, Óskarsdóttir M (2021) The future of sleep measurements: a review and perspective. Sleep Med Clin 16(3):447–464
Arnardottir ES, Islind AS, Óskarsdóttir M et al (2022) The sleep revolution project: the concept and objectives. J Sleep Res 31(4):e13,630
Bagnall A, Lines J, Bostrom A et al (2017) The great time series classification bake off: a review and experimental evaluation of recent algorithmic advances. Data Mini Knowl Discov 31(3):606–660
Biedebach L, Rusanen M, Leppänen T et al (2023) Towards a deeper understanding of sleep stages through their representation in the latent space of variational autoencoders. In: proceedings of the annual Hawaii international conference on system sciences, IEEE Computer Society, pp 3111–3120
Blázquez-García A, Conde A, Mori U et al (2021) A review on outlier/anomaly detection in time series data. ACM Comput Surv (CSUR) 54(3):1–33
Chalapathy R, Chawla S (2019) Deep learning for anomaly detection: A survey. ar**v preprint ar**v:1901.03407
Chandola V, Banerjee A, Kumar V (2009) Anomaly detection: a survey. ACM Comput Surv (CSUR) 41(3):1–58
Chauhan S, Vig L (2015) Anomaly detection in ecg time signals via deep long short-term memory networks. In: 2015 IEEE international conference on data science and advanced analytics (DSAA), IEEE, pp 1–7
Curran K, Yuan P, Coyle D (2012) Using acoustic sensors to discriminate between nasal and mouth breathing. Int J Bioinform Res Appl 8(5–6):382–396
Danker-hopfe H, Anderer P, Zeitlhofer J et al (2009) Interrater reliability for sleep scoring according to the rechtschaffen & kales and the new aasm standard. J Sleep Res 18(1):74–84
de Castilho LS, Abreu MHNG, de Oliveira RB et al (2016) Factors associated with mouth breathing in children with developmental disabilities. Spec Care Dent 36(2):75–79
Deng H, Runger G, Tuv E et al (2013) A time series forest for classification and feature extraction. Inform Sci 239:142–153
Denotti G, Ventura S, Arena O et al (2014) Oral breathing: new early treatment protocol. J Pediat Neonat Individ Med (JPNIM) 3(1):e030,108-e030,108
Fawaz HI, Forestier G, Weber J et al (2019) Deep learning for time series classification: a review. Data Min Knowl Disc 33(4):917–963
Fensterseifer GS, Carpes O, Weckx LLM et al (2013) Mouth breathing in children with learning disorders. Braz J Otorhinolaryngol 79:620–624
Fleming S, Thompson M, Stevens R et al (2011) Normal ranges of heart rate and respiratory rate in children from birth to 18 years of age: a systematic review of observational studies. The Lancet 377(9770):1011–1018
Forman G, Scholz M (2010) Apples-to-apples in cross-validation studies: pitfalls in classifier performance measurement. ACM Sigkdd Explorat Newsl 12(1):49–57
Freeman C, Merriman J, Beaver I et al (2021) Experimental comparison and survey of twelve time series anomaly detection algorithms. J Artif Intell Res 72:849–899
Fu K, Cheng D, Tu Y, et al (2016) Credit card fraud detection using convolutional neural networks. In: neural information processing: 23rd international conference, ICONIP 2016, Kyoto, Japan, October 16–21, 2016, Proceedings, Part III 23, Springer, pp 483–490
Gao J, Murphey YL, Zhu H (2018) Multivariate time series prediction of lane changing behavior using deep neural network. Appl Intell 48(10):3523–3537
Gozal D (1998) Sleep-disordered breathing and school performance in children. Pediatrics 102(3):616–620
Grabenhenrich L, Trendelenburg V, Bellach J et al (2020) Frequency of food allergy in school-aged children in eight European countries-the Europrevall-Ifaam birth cohort. Allergy 75(9):2294–2308
Gunnarsson BR, Vanden Broucke S, Baesens B et al (2021) Deep learning for credit scoring: do or don’t? Europ J Operat Res 295(1):292–305
Henry KE, Kornfield R, Sridharan A et al (2022) Human-machine teaming is key to ai adoption: clinicians’ experiences with a deployed machine learning system. NPJ Dig Med 5(1):97
Holm B, Óttir M, Arnardóttir ES, et al (2022) Automatic non-invasive isolation of respiratory cycles. ar**v preprint ar**v:2203.01828
Huang G, Ma F (2021) Concad: contrastive learning-based cross attention for sleep apnea detection. In: joint european conference on machine learning and knowledge discovery in databases, Springer, pp 68–84
Hudgel DW, Martin RJ, Johnson B et al (1984) Mechanics of the respiratory system and breathing pattern during sleep in normal humans. J Appl Physiol 56(1):133–137
Izu SC, Itamoto CH, Pradella-Hallinan M et al (2010) Obstructive sleep apnea syndrome (Osas) in mouth breathing children. Braz J Otorhinolaryngol 76:552–556
Kainulainen S, Korkalainen H, Sigurdardóttir S et al (2021) Comparison of eeg signal characteristics between polysomnography and self applied somnography setup in a pediatric cohort. IEEE Access 9:110,916-110,926
Keil T, McBride D, Grimshaw K et al (2010) The multinational birth cohort of Europrevall: background, aims and methods. Allergy 65(4):482–490
Kemp B, Värri A, Rosa AC et al (1992) A simple format for exchange of digitized polygraphic recordings. Electroencephal Clin Neurophysiol 82(5):391–393
Korkalainen H, Aakko J, Nikkonen S et al (2019) Accurate deep learning-based sleep staging in a clinical population with suspected obstructive sleep apnea. IEEE J Biomed Health Inform 24(7):2073–2081
Koutsourelakis I, Vagiakis E, Roussos C et al (2006) Obstructive sleep Apnoea and oral breathing in patients free of nasal obstruction. Europ Respir J 28(6):1222–1228
Le Nguyen T, Gsponer S, Ilie I et al (2019) Interpretable time series classification using linear models and multi-resolution multi-domain symbolic representations. Data Min Knowl Discov 33(4):1183–1222
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Lee SY, Guilleminault C, Chiu HY et al (2015) Mouth breathing, nasal disuse, and pediatric sleep-disordered breathing. Sleep Breath 19(4):1257–1264
Lemnaru C, Potolea R (2011) Imbalanced classification problems: systematic study, issues and best practices. In: international conference on enterprise information systems, Springer, pp 35–50
Leung K, Leckie C (2005) Unsupervised anomaly detection in network intrusion detection using clusters. Proc Twenty-Eighth Austral Conf Comput Sci 38:333–342
Li L, Yan J, Wang H et al (2020) Anomaly detection of time series with smoothness-inducing sequential variational auto-encoder. IEEE Trans Neural Netw Learn Syst 32(3):1177–1191
Malhotra P, Vig L, Shroff G, et al (2015) Long short term memory networks for anomaly detection in time series. In: Proceedings, pp 89–94
Marcus CL (2001) Sleep-disordered breathing in children. Am J Respirat Crit Care Med 164(1):16–30
Markun LC, Sampat A (2020) Clinician-focused overview and developments in polysomnography. Curr Sleep Med Rep 6:309
Natekin A, Knoll A (2013) Gradient boosting machines, a tutorial. Front Neurorob 7:21
Oner MU, Cheng YC, Lee HK, et al (2020) Training machine learning models on patient level data segregation is crucial in practical clinical applications. medRxiv 2020–04
Park D, Hoshi Y, Kemp CC (2018) A multimodal anomaly detector for robot-assisted feeding using an lstm-based variational autoencoder. IEEE Robot Autom Lett 3(3):1544–1551
Peralta M, Jannin P, Baxter JS (2021) Machine learning in deep brain stimulation: A systematic review. Artificial Intelligence in Medicine 122(102):198
Ratanamahatana CA, Keogh E (2005) Three myths about dynamic time war** data mining. In: proceedings of the 2005 SIAM international conference on data mining, SIAM, 506–510
Rewicki F, Denzler J, Niebling J (2023) Is it worth it? Comparing six deep and classical methods for unsupervised anomaly detection in time series. Appl Sci 13(3):1778
Ribeiro M, Lazzaretti AE, Lopes HS (2018) A study of deep convolutional auto-encoders for anomaly detection in videos. Patt Recogn Lett 105:13–22
Sabil A, Glos M, Günther A et al (2019) Comparison of apnea detection using oronasal thermal airflow sensor, nasal pressure transducer, respiratory inductance plethysmography and tracheal sound sensor. J Clin Sleep Med 15(2):285–292
Sano M, Sano S, Kato H et al (2018) Proposal for a screening questionnaire for detecting habitual mouth breathing, based on a mouth-breathing habit score. BMC Oral Health 18(1):1–13
Shwartz-Ziv R, Armon A (2022) Tabular data: Deep learning is not all you need. Information Fusion 81:84–90
Sigurdardóttir ST, Jonasson K, Clausen M et al (2021) Prevalence and early-life risk factors of school-age allergic multimorbidity: the europrevall-ifaam birth cohort. Allergy 76(9):2855–2865
Zhao B, Lu H, Chen S et al (2017) Convolutional neural networks for time series classification. J Syst Eng Electron 28(1):162–169
Acknowledgements
We would like to thank sleep technologist Kristín Anna Ólafsdóttir who was part of the team that set up the PSG devices for the children. Furthermore, we would like to thank the children and their parents for their contribution to this study. This project received funding from the European Unions Horizon 2020 research and innovation program under grant agreement No. 965417. The project was supported financially by the Icelandic Research Fund 2016-2019, No. 174067, Nordforsk 2018-2021, (NordSleep, No. 90458) and the Landspitali University Hospital Science Fund 2019-2020 (No. 893831). Nox Medical (Reykjavik, Iceland) additionally supported the study by supplying the researchers with the A1 sleep recorders and consumables needed for the sleep studies. The birth cohort study was funded by the European Commission: (a) under the 6th Framework Programme (FOOD-CT-2005-514000) within the collaborative research initiative ‘EuroPrevall’, and (b) under the 7th Framework Programme (FP7-KBBE-2012-6; grant agreement No. 312 147) within the collaborative project ‘iFAAM’.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Johannes Fürnkranz.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Biedebach, L., Óskarsdóttir, M., Arnardóttir, E.S. et al. Anomaly detection in sleep: detecting mouth breathing in children. Data Min Knowl Disc 38, 976–1005 (2024). https://doi.org/10.1007/s10618-023-00985-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10618-023-00985-x