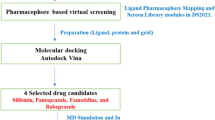
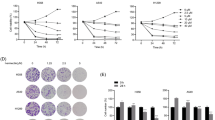
Abstract
The anillin actin-binding protein (ANLN) is immensely overexpressed in cancers, including lung cancer (LC). Phytocompounds have gained interest due to their broader potential and reduced unwanted effects. Screening numerous compounds presents a challenge, but in silico molecular docking is pragmatic. The present study aims to identify the role of ANLN in lung adenocarcinoma (LUAD), along with identification and interaction analysis of anticancer and ANLN inhibitory phytocompounds followed by molecular dynamics (MD) simulation. Using a systematic approach, we found that ANLN is significantly overexpressed in LUAD and mutated with a frequency of 3.73%. It is linked with advanced stages, clinicopathological parameters, worsening of relapse-free survival (RFS), and overall survival (OS), pinpointing its oncogenic and prognostic potential. High-throughput screening and molecular docking of phytocompounds revealed that kaempferol (flavonoid aglycone) interacts strongly with the active site of ANLN protein via hydrogen bonds, Vander Waals interactions, and acts as a potent inhibitor. Furthermore, we discovered that ANLN expression was found to be significantly higher (p) in LC cells compared to normal cells. This is a propitious and first study to demonstrate ANLN and kaempferol interactions, which might eventually lead to removal of rout from cell cycle regulation posed by ANLN overexpression and allow it to resume normal processes of proliferation. Overall, this approach suggested a plausible biomarker role of ANLN and the combination of molecular docking subsequently led to the identification of contemporary phytocompounds, bearing symbolic anticancer effects. The findings would be advantageous for pharmaceutics but require validation using in vitro and in vivo methods.
Highlights
• ANLN is significantly overexpressed in LUAD.
• ANLN is implicated in the infiltration of TAMs and altering plasticity of TME.
• Kaempferol (potential ANLN inhibitor) shows important interactions with ANLN which could remove the alterations in cell cycle regulation, imposed by ANLN overexpression eventually leading to normal process of cell proliferation.






Similar content being viewed by others
Data availability
No data was used for the research described in the article.
Change history
13 July 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10142-023-01174-1
Abbreviations
- LC:
-
Lung cancer
- NSCLC:
-
Non-small cell lung cancer
- LUAD:
-
Lung adenocarcinoma
- mRNA:
-
Messenger RNA
- NCBI:
-
National Center for Biotechnology Information
- ANLN:
-
Anillin
- DEGs:
-
Differentially expressed genes
- OS:
-
Overall survival
- KM:
-
Kaplan-Meier
- LUSC:
-
Lung squamous cell carcinoma
- RhoA:
-
Ras homolog family member A
- APC/C:
-
Anaphase-promoting complex/cyclosome
- BLCA:
-
Bladder urothelial carcinoma
- ATC:
-
Anaplastic thyroid carcinoma
- NPC:
-
Nasopharyngeal carcinoma
- RCC:
-
Renal cell carcinoma
- RFS:
-
Recurrence-free survival
- TME:
-
Tumor microenvironment
- MD:
-
Molecular dynamics
- GEPIA:
-
Gene expression profiling interactive analysis
- GO:
-
Gene ontology
- BP:
-
Biological process
- MF:
-
Molecular function
- CC:
-
Cellular compartment
- TCGA:
-
The Cancer Genome Atlas
- TIMER:
-
Tumor immune estimation resource
- 3D:
-
Three-dimensional
- RCSB PDB:
-
Research Collaboratory for Structural Bioinformatics Protein Data Bank
- ADME:
-
Absorption distribution metabolism and excretion
- DS:
-
Discovery Studio
- 2D:
-
Two-dimensional
- GAFF:
-
General amber force field
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- MM/PBSA:
-
Molecular mechanics/Poisson-Boltzmann surface area
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- FBS:
-
Fetal bovine serum
- cDNA:
-
Complementary DNA
- PCR:
-
Polymerase chain reaction
- qRT-PCR:
-
Real-time quantitative reverse transcription PCR
- HPA:
-
Human Protein Atlas
- COAD:
-
Colon adenocarcinoma
- BRCA:
-
Breast invasive carcinoma
- STAD:
-
Stomach adenocarcinoma
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- DLBC:
-
Lymphoid neoplasm diffuse large B cell lymphoma
- ESCA:
-
Esophageal carcinoma
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- THYM:
-
Thymoma
- UCEC:
-
Uterine corpus endometrial carcinoma
- UCS:
-
Uterine carcinosarcoma
- KIF23:
-
Kinesin family member 23
- C16orf89:
-
Chromosome 16 open reading frame 89
- PLK4:
-
Polo-like kinase 4
- BTG2:
-
BTG anti-proliferation factor 2
- TP53:
-
Tumor protein P53
- NKT:
-
Natural killer T
- SMILES:
-
Simplified molecular-input line-entry system
- MW:
-
Molecular weight
- H-bonds:
-
Hydrogen bonds
- \({{{R}}}_{{g}}\) :
-
Radius of gyration
- GAP:
-
GTPase-activating protein
- GEF:
-
Rho guanine nucleotide exchange factor
- KIF4A:
-
Kinesin family member 4A
- TPX2:
-
TPX2 microtubule nucleation factor
- DLGAP5:
-
DLG-associated protein 5
- GKAP:
-
Guanylate-kinase-associated protein
- TAMs:
-
Tumor-associated macrophages
- PME:
-
Particle-mesh Ewald algorithm
References
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Arora S, Singh P, Rahmani AH, Almatroodi SA, Dohare R, Syed MA (2020) Unravelling the role of miR-20b-5p, CCNB1, HMGA2 and E2F7 in development and progression of non-small cell lung cancer (NSCLC). Biology 9:201. https://doi.org/10.3390/biology9080201
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. https://doi.org/10.1158/2159-8290.CD-12-0095
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S (2017) UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19:649–658. https://doi.org/10.1016/j.neo.2017.05.002
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Govindaraju S, Roshini A, Lee M-H, Yun K (2019) Kaempferol conjugated gold nanoclusters enabled efficient for anticancer therapeutics to A549 lung cancer cells. IJN 14:5147–5157. https://doi.org/10.2147/IJN.S209773
Győrffy B, Surowiak P, Budczies J, Lánczky A (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 8:e82241. https://doi.org/10.1371/journal.pone.0082241
Herbst RS, Heymach JV, Lippman SM (2008) Lung Cancer. N Engl J Med 359:1367–1380. https://doi.org/10.1056/NEJMra0802714
Hickson GRX, O’Farrell PH (2008) Rho-dependent control of anillin behavior during cytokinesis. J Cell Biol 180:285–294. https://doi.org/10.1083/jcb.200709005
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725. https://doi.org/10.1002/prot.21123
Idichi T, Seki N, Kurahara H, Yonemori K, Osako Y, Arai T, Okato A, Kita Y, Arigami T, Mataki Y, Kijima Y, Maemura K, Natsugoe S (2017) Regulation of actin-binding protein ANLN by antitumor miR-217 inhibits cancer cell aggressiveness in pancreatic ductal adenocarcinoma. Oncotarget 8:53180–53193. https://doi.org/10.18632/oncotarget.18261
Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA (2019) MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med 70:3–20. https://doi.org/10.1016/j.mam.2018.07.003
Jakalian A, Bush BL, Jack DB, Bayly CI (2000) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method J Comput Chem 21:132–146. https://doi.org/10.1002/(SICI)1096-987X(20000130)21:2%3c132::AID-JCC5%3e3.0.CO;2-P
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23:1623–1641. https://doi.org/10.1002/jcc.10128
Jonnalagadda B, Arockiasamy S, Vetrivel U, PA A, (2020) In silico docking of phytocompounds to identify potent inhibitors of signaling pathways involved in prostate cancer. J Biomol Struct Dyn 39(14):5182–5208. https://doi.org/10.1080/07391102.2020.1785944
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897. https://doi.org/10.1021/ar000033j
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan, A., (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90-97. https://doi.org/10.1093/nar/gkw377
Kumari R, Kumar R, Open source drug discovery consortium, Lynn A (2014) g_mmpbsa —a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. https://doi.org/10.1021/ci500020m
Lagunin A, Stepanchikova A, Filimonov D, Poroikov V (2000) PASS: prediction of activity spectra for biologically active substances. Bioinformatics 16:747–748. https://doi.org/10.1093/bioinformatics/16.8.747
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48:W509–W514. https://doi.org/10.1093/nar/gkaa407
Liang J, Cui Y, Meng Y, Li X, Wang X, Liu W, Huang L, Du H (2019) Integrated analysis of transcription factors and targets co-expression profiles reveals reduced correlation between transcription factors and target genes in cancer. Funct Integr Genomics 19:191–204. https://doi.org/10.1007/s10142-018-0636-6
Long X, Zhou W, Wang Y, Liu S (2018) Prognostic significance of ANLN in lung adenocarcinoma. Oncol Lett. https://doi.org/10.3892/ol.2018.8858
Magnusson K, Gremel G, Rydén L, Pontén V, Uhlén M, Dimberg A, Jirström K, Pontén F (2016) ANLN is a prognostic biomarker independent of Ki-67 and essential for cell cycle progression in primary breast cancer. BMC Cancer 16:904. https://doi.org/10.1186/s12885-016-2923-8
Mammalian Gene Collection (MGC) Program Team (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci 99:16899–16903. https://doi.org/10.1073/pnas.242603899
Mohammad T, Mathur Y, Hassan MI (2020) InstaDock: a single-click graphical user interface for molecular docking-based virtual high-throughput screening. Brief Bioinform 22(4):bbaa279. https://doi.org/10.1093/bib/bbaa279
Oegema K, Savoian MS, Mitchison TJ, Field CM (2000) Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol 150:539–552. https://doi.org/10.1083/jcb.150.3.539
Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase T, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, Sugano S (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 36:40–45. https://doi.org/10.1038/ng1285
Ozer B, Sezerman U (2017) Analysis of the interplay between methylation and expression reveals its potential role in cancer aetiology. Funct Integr Genomics 17:53–68. https://doi.org/10.1007/s10142-016-0533-9
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1–6. https://doi.org/10.1016/s1476-5586(04)80047-2
Rigsby RE, Parker AB (2016) Using the PyMOL application to reinforce visual understanding of protein structure: PyMOL Application to Understand Protein Structure. Biochem Mol Biol Educ 44:433–437. https://doi.org/10.1002/bmb.20966
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Shriwash N, Singh P, Arora S, Ali SM, Ali S, Dohare R (2019) Identification of differentially expressed genes in small and non-small cell lung cancer based on meta-analysis of mRNA. Heliyon 5:e01707. https://doi.org/10.1016/j.heliyon.2019.e01707
Singh P, Gurung R, Sultan A, Dohare R (2023) Understanding the role of adipokines and adipogenesis family in hepatocellular carcinoma. Egypt J Med Hum Genet 24:17. https://doi.org/10.1186/s43042-023-00401-5
Sjöstedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, Oksvold P, Edfors F, Limiszewska A, Hikmet F, Huang J, Du Y, Lin L, Dong Z, Yang L, Liu X, Jiang H, Xu X, Wang J, Yang H, Bolund L, Mardinoglu A, Zhang C, von Feilitzen K, Lindskog C, Pontén F, Luo Y, Hökfelt T, Uhlén M, Mulder J (2020) An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367:eaay5947. https://doi.org/10.1126/science.aay5947
Solanki R, Singh P, Beg MA, Dohare R, Verma AK, Ahmad FJ, Alankar B, Athar F, Kaur H (2023) Combined transcriptomics and in-silico approach uncovers the role of prognostic biomarkers in hepatocellular carcinoma. Human Gene 35:201154. https://doi.org/10.1016/j.humgen.2023.201154
Sousa da Silva AW, Vranken WF (2012) ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res Notes 5:367. https://doi.org/10.1186/1756-0500-5-367
Straight AF, Field CM, Mitchison TJ (2005) Anillin binds nonmuscle myosin II and regulates the contractile ring. MBoC 16:193–201. https://doi.org/10.1091/mbc.e04-08-0758
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47:W556–W560. https://doi.org/10.1093/nar/gkz430
Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, Bäckström A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson Å, Sjöstedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Pontén F, von Feilitzen K, Lilley KS, Uhlén M, Lundberg E (2017) A subcellular map of the human proteome. Science 356:eaal3321. https://doi.org/10.1126/science.aal3321
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F (2015) Tissue-based map of the human proteome. Science 347:1260419–1260419. https://doi.org/10.1126/science.1260419
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnström H, Glimelius B, Sjöblom T, Edqvist P-H, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F (2017) A pathology atlas of the human cancer transcriptome. Science 357:eaan2507. https://doi.org/10.1126/science.aan2507
Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, Lakshmikanth T, Forsström B, Edfors F, Odeberg J, Mardinoglu A, Zhang C, von Feilitzen K, Mulder J, Sjöstedt E, Hober A, Oksvold P, Zwahlen M, Ponten F, Lindskog C, Sivertsson Å, Fagerberg L, Brodin P (2019) A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 366:eaax9198. https://doi.org/10.1126/science.aax9198
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Wang A, Dai H, Gong Y, Zhang C, Shu J, Luo Y, Jiang Y, Liu W, Bie P (2019) ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis. J Exp Clin Cancer Res 38:347. https://doi.org/10.1186/s13046-019-1340-7
Xu J, Zheng H, Yuan S, Zhou B, Zhao W, Pan Y, Qi D (2019) Overexpression of ANLN in lung adenocarcinoma is associated with metastasis. Thorac Cancer 10:1702–1709. https://doi.org/10.1111/1759-7714.13135
Zeng S, Yu X, Ma C, Song R, Zhang Z, Zi X, Chen X, Wang Y, Yu Y, Zhao J, Wei R, Sun Y, Xu C (2017) Transcriptome sequencing identifies ANLN as a promising prognostic biomarker in bladder urothelial carcinoma. Sci Rep 7:3151. https://doi.org/10.1038/s41598-017-02990-9
Zheng X, Ma H, Dong Y, Fang M, Wang J, **ong X, Liang J, Han M, You A, Yin Q, Huang W (2023) Immune-related biomarkers predict the prognosis and immune response of breast cancer based on bioinformatic analysis and machine learning. Funct Integr Genomics 23:201. https://doi.org/10.1007/s10142-023-01124-x
Acknowledgements
We would like to thank Jamia Millia Islamia, University of Lucknow, ICGEB, AIIMS for providing infrastructure, internet facility, and journal access. Prithvi Singh would like to thank the Indian Council of Medical Research (ICMR), Government of India, for awarding him a Senior Research Fellowship [Grant Number: BMI/11(89)/2020].
Funding
This research was funded by the ICMR Extramural grant awarded to Mansoor Ali Syed (Grant no: 5/30/59/2020/NCD3).
Author information
Authors and Affiliations
Contributions
Prithvi Singh: Conceptualization, Methodology, Software, Formal analysis, Investigation, Visualization, Data curation, Writing—Original Draft, Writing—Review and Editing. Shweta Arora: Formal analysis, Data curation, Writing-Original Draft, Writing—Review and Editing. Md Amjad Beg: Methodology, Software, Data curation, Writing-Original Draft, Writing-Review and Editing. Sibasis Sahoo: Methodology, Software, Data curation, Writing-Original Draft, Writing—Review and Editing. Arnab Nayek: Methodology, Software, Data curation, Writing-Original Draft, Writing—Review and Editing. Mohd Mabood Khan: Methodology, Software, Visualization, Validation, Investigation, Writing-Original Draft, Writing—Review and Editing. Anuradha Sinha: Methodology, Validation, Visualization, Writing—Original Draft, Writing—Review and Editing. Md. Zubbair Malik: Writing—Original Draft, Writing-Review and Editing. Fareeda Athar: Writing—Original Draft, Writing—Review and Editing. Mohammad Serajuddin: Writing-Original Draft, Writing—Review and Editing. Ravins Dohare: Writing—Review and Editing, Supervision, Project Administration. Mansoor Ali Syed: Resources, Writing—Review and Editing, Supervision, Project Administration. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Originally, the online published article contains an error. The two complete author names should be “Md Amjad Beg” and “Md. Zubbair Malik”. Md shoud be part of the author’s given name.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, P., Arora, S., Beg, M.A. et al. Comprehensive multiomics and in silico approach uncovers prognostic, immunological, and therapeutic roles of ANLN in lung adenocarcinoma. Funct Integr Genomics 23, 223 (2023). https://doi.org/10.1007/s10142-023-01144-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-01144-7