Abstract
Birds are important hosts for many RNA viruses, including influenza A virus, Newcastle disease virus, West Nile virus and coronaviruses. Innate defense against RNA viruses in birds involves detection of viral RNA by pattern recognition receptors. Several receptors of different classes are involved, such as endosomal toll-like receptors and cytoplasmic retinoic acid–inducible gene I-like receptors, and their downstream adaptor proteins. The function of these receptors and their antagonism by viruses is well established in mammals; however, this has received less attention in birds. These receptors have been characterized in a few bird species, and the completion of avian genomes will permit study of their evolution. For each receptor, functional work has established ligand specificity and activation by viral infection. Engagement of adaptors, regulation by modulators and the supramolecular organization of proteins required for activation are incompletely understood in both mammals and birds. These receptors bind conserved nucleic acid agonists such as single- or double-stranded RNA and generally show purifying selection, particularly the ligand binding regions. However, in birds, these receptors and adaptors differ between species, and between individuals, suggesting that they are under selection for diversification over time. Avian receptors and signalling pathways, like their mammalian counterparts, are targets for antagonism by a variety of viruses, intent on escape from innate immune responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Birds are important hosts of many RNA viruses, some having zoonotic potential. Ducks and shorebirds are critical reservoir hosts of influenza A viruses, propagating all strains (Olsen et al. 2006; Webster et al. 1992). West Nile virus cycles between birds and mosquitoes, with humans and horses as accidental infections (Saiz et al. 2021). More than 30 different avian species can be experimentally infected, implicating them in transmission (Komar et al. 2003). Newcastle disease viruses (NDV) are capable of infecting over 200 avian species, and changes in virulence due to cross-species adaptation have been thoroughly reviewed (Afonso 2021). Wild aquatic birds are hosts to a diversity of coronaviruses (Chu et al. 2011). Recently, virome analyses in waterfowl and shorebirds identified 27 viruses, 24 of which were novel (Wille et al. 2019). These studies reveal multi-host generalist viruses (like influenza and coronaviruses) and others that are host-specific. Our knowledge of avian RNA viruses is far from complete, and prediction of zoonotic risk is difficult (Wille et al. 2021). RNA viruses are constantly evolving in these hosts and have diverse strategies to antagonize host immunity.
Innate immunity provides the first line of defense against microbial pathogens. Recognition stimulates innate immune signalling leading to synthesis of a host of antimicrobial effectors, as well as instructing the adaptive immune response to the type of response to make (Iwasaki and Medzhitov 2015). The initial step of detection and recognition is key to initiation of a successful immune response (Janeway 1989). Pathogen recognition involves pattern recognition receptors on the cell surface and within the cytoplasm. These pattern recognition receptors (PRR) recognize pathogen parts or pathogen associated molecular patterns (PAMPs), typically very conserved molecular structures, which are often also recognized as danger associated molecular patterns (DAMPs) arising from within the cell. Pattern recognition of RNA viruses involves primarily toll-like receptors (Kawai and Akira 2010) and intracellular nucleic acid detectors (Hur 2019; Wu and Chen 2014).
Avian pattern recognition receptors have been reviewed recently (Neerukonda and Katneni 2020), and avian sensing of viral pathogens reviewed in 2013 (Chen et al. 2013). Two reviews recently examine avian detection and response to influenza (Campbell and Magor 2020; Evseev and Magor 2019) and avian toll-like receptor (TLR) expression and variants in association with response to various pathogens (Rehman et al. 2021). In the last 10 years, much has been learned about avian pattern recognition receptors. Furthermore, comparisons of receptors in related species emerging from genome projects reveal interesting patterns of evolution of these receptors in avian species (Velova et al. 2018). Here, I will explore the function and evolution of pattern recognition receptors of birds involved in the detection of RNA viruses through detection of nucleic acids and their signalling pathways. The main signalling pathways downstream of RNA detection converge on inducing type I interferons and proinflammatory cytokines (Fig. 1). Functional data from avian PRRs and signalling components (Table 1) support the pathways shown and molecular details are predicted to be conserved across species.
RNA viruses are primarily detected in avian cells by binding of RNA by endosomal TLRs and cytoplasmic nucleic acid sensors. Single-stranded RNA is detected by TLR7 (primarily in B cells and possibly pDCs). Double-stranded RNA is detected by TLR3 and MDA5. RIG-I detects 5′-triphosphate RNA panhandle structures. E3 ubiquitin ligases are involved in activation of mammalian RIG-I, and MDA5; however, this is still unclear in birds. Both RIG-I and RIPLET are missing in chickens. STING potentiates the signal from MDA5 and also detects viral fusion. Oligomerization of components (MAVS) or formation of a supramolecular complex (myddosome or triffosome) initiates the signalling. Phosphorylation of adaptor proteins TRIF, MAVS, and STING by TBK1 (or IKK) recruits IRF7 (phosphorylation in red), whereby it becomes phosphorylated by TBK1 (blue) and dimerizes to enter the nucleus and drive transcription of type I interferons. Signalling from MyD88 recruits IRAK and TRAF6 to activate IRF7 and degrade the inhibitor of NF-κB to drive transcription of proinflammatory cytokines. Intracellular signalling is inferred from interpretation of functional data and conserved signalling mechanisms, many of which still need to be experimentally tested in birds.
Toll-like receptors and supramolecular organization
Toll-like receptors (TLR) are mediators of detection and signalling in response to conserved molecular patterns and were recently the subject of a comprehensive and excellent review (Fitzgerald and Kagan 2020). TLRs form a multigene family that are found in vertebrates and invertebrates, composed of 10 members in humans (Kawai and Akira 2006a) and 10 in chickens (Boyd et al. 2007; Brownlie and Allan 2011; Temperley et al. 2008). TLR are composed of leucine-rich repeats which form the agonist binding region, and an intracellular toll/interleukin-1 receptor (TIR) domain which connects to downstream adaptors for signalling (Kawai and Akira 2006b). TLRs dimerize when stimulated by the detection of infectious agents, and their signalling triggers the generation of the supramolecular organizing centers involved in co-ordinating the cellular signals through transcription factors and metabolic changes (Fitzgerald and Kagan 2020). TLR signalling is divided by the adaptors recruited, with myeloid differentiation primary response 88 (MyD88) leading to proinflammatory cytokine signalling, or TIR domain containing adaptor (TRIF) (Hoebe et al. 2003; Yamamoto et al. 2003) leading to interferon. All TLRs except TLR3, interact with TIRAP/Mal and MyD88, generating a multiprotein complex termed the myddosome (Motshwene et al. 2009), while TLR3 and TLR4 interact with TRAM and TRIF forming an analogous signalling platform called a triffosome (Fitzgerald and Kagan 2020). The MyD88 myddosome recruits TRAF6 which activates TBK1. The formation of the supramolecular complex and oligomerization of additional components are necessary for activation of E3-ubiquitin ligase TRAF6 and the associated kinase transforming growth factor β (TGFβ)–activated kinase 1(TAK1) (Wang et al. 2001). This kinase complex phosphorylates IκBα within the inhibitory IκB complex which tags it for polyubiquitination and subsequent degradation. Release of the inhibitor uncovers a nuclear localization signal on nuclear factor kappa B (NFκB) heterodimers (Karin 1999), which translocate to the nucleus and bind target sites in the promoters and enhancers of immune genes including TNF-alpha and other proinflammatory cytokines.
On the endosomal membrane, TLR4 interacts with TRAM and TRIF, and TLR3 also recruits TRIF, both leading to activation of TANK binding kinase (TBK1) which activates IRF3 and drives transcription of interferon and interferon stimulated genes (Fitzgerald et al. 2003; McWhirter et al. 2004; Sharma et al. 2003). TLR3 uses only TRIF for interferon induction and is the only TLR that does not signal through MyD88. TRIF signals through TBK1 and recruits interferon regulatory factor 3 (IRF3), leading to the production of type I interferon. The phosphorylation site in TRIF resides in a consensus pLxIS motif, which is shared with other intracellular adaptors (Liu et al. 2015b). In this motif, the p is any hydrophilic residue, L leucine, x any residue and phosphorylation site at S serine.
Many of the components of PRR signaling, including MyD88, TRIF and TBK1 are conserved in birds (Cormican et al. 2009; Gillespie et al. 2011; Wang et al. 2017; Wheaton et al. 2007); however, IRF3 is notably absent. In mammals, IRF3 is constitutively expressed and dimerizes upon phosphorylation to enter the nucleus and turn on type I interferon (Lin et al. 1998). Interferon activates IRF7, a related regulatory factor, that is critically involved in both TLR and RIG-I-like receptor (RLR) signalling (Honda et al. 2005). A gene was originally named IRF3 in chickens (Grant et al. 1995); however, it is interferon-inducible and more closely resembles IRF7 in sequence and synteny of surrounding genes (Santhakumar et al. 2017). Others have previously noted the absence of IRF3 in birds (Cormican et al. 2009; Huang et al. 2010). Chicken IRF7 translocates to the nucleus and drives transcription of an interferon reporter (**ao et al. 2018), and overexpression of duck IRF7 decreases viral replication (Chen et al. 2019). The absence of IRF3 and key role of chIRF7 were confirmed by analyzing de novo assembled RNA-seq data in response to the dsRNA viral mimic poly (I:C), following IRF7 knockdown or overexpression (Kim and Zhou 2015). Ultimately, knockout of IRF7 in the DF-1 chicken embryonic fibroblast cell line (Cheng et al. 2019b; Kim et al. 2020) greatly decreased expression of type I interferon. In birds, two serologically distinct types of type I interferon genes have been identified on the Z chromosome (Sick et al. 1996). Of these, there are at least 10 genes with a core region resembling IFN-alpha (IFN-α), while chicken IFN-2 has similarities to IFN-beta (IFN-β). Their presumed receptor composed of IFNAR1 and IFNAR2 has been identified in chickens (Guo et al. 2014). Interferon signalling leads to the induction of interferon stimulated genes (ISGs) many of which have antiviral function, as reviewed recently (Campbell and Magor 2020; Evseev and Magor 2019; Santhakumar et al. 2017).
Avian TLR receptors
The composition of avian TLR genes has been the subject of several comprehensive reviews (Boyd et al. 2007; Brownlie and Allan 2011; Cormican et al. 2009; Keestra et al. 2013; Nawab et al. 2019; Temperley et al. 2008; Velova et al. 2018). Birds have 10 TLR genes with most being direct orthologues, while others are unique to birds. Two genes representing TLR1 (types A and B), two TLR2 (types A and B), TLR3, TLR4, TLR5 and TLR7 appear to be orthologous genes. Phylogenetic relationships of TLR1 and TLR2 to mammalian orthologues have been inferred (Huang et al. 2011). Inclusion of more species and construction of phylogenetic trees based on the regions excluded from gene conversion events, support their conclusions and suggest that TLR1B should be renamed TLR1, while TLR1A is the TLR10 orthologue (Velova et al. 2018). With emerging avian genomes, recent phylogenetic analyses of TLRs from 50 bird species show a complicated history of duplications of TLR7 in some species, deletions of TLR8 and pseudogenization of TLR5 in some birds (Velova et al. 2018). TLR15 is unique to birds and reptiles (Boyd et al. 2012; Higgs et al. 2006), and TLR21 is also found in fish and amphibians (Brownlie et al. 2009; Keestra et al. 2010).
Similar to their mammalian counterparts, each avian TLR orthologue has specificity for different pathogen molecular patterns. TLR1 and 2 have specificity for lipoproteins and peptidoglycans (Higgs et al. 2006; Yilmaz et al. 2005), TLR3 for double-stranded RNA (Karpala et al. 2008), TLR4 for lipopolysaccharide (Keestra and van Putten 2008), TLR5 for flagellin monomers (Iqbal et al. 2005b) and TLR7 binds single-stranded RNA (Philbin et al. 2005). TLR15 has specificity for a protein component of yeast that must be enzymatically processed (Boyd et al. 2012) and diacylated lipopeptide from mycoplasma (Oven et al. 2013). Alternatively, TLR15 itself is proteolytically cleaved by microbial proteases for activation (de Zoete et al. 2011). TLR21 is the avian sensor for CpG, analogous to TLR9 (Brownlie et al. 2009). MyD88 and TRIF adaptors are present in birds, but TRAM, the TLR4 associated adaptor used for signalling interferon, is not (Keestra et al. 2013). The chicken macrophage–like cell line HD-11 does not activate IFNβ in response to stimulation with LPS (Keestra and van Putten 2008).
TLR function is restricted by cellular location, although this differs with cell type. Mammalian TLR1, TLR2, TLR4 and TLR5 are present on cellular membranes where they survey the extracellular milieu (Kawai and Akira 2006b), and the chicken orthologs are presumed to be on the cell surface (Neerukonda and Katneni 2020). Avian TLR15 is also on the cell surface (Boyd et al. 2012). Avian TLR3 and TLR7 are present on the endosomal membrane where they sample the contents of the endosome. TLR3 and TLR7 are the most important for detection of viruses internalized through receptor mediated endocytosis, as their nucleic acid can be detected by the TLRs in the endosome (Lund et al. 2004). Similarly, TLR21 localizes to the endosomal membrane for detection of CpG DNA from bacterial or viral DNA (Brownlie et al. 2009). The restriction of these TLRs to the endosomal compartment ensures that the entry of nucleic acids from incoming viral particles can be detected, while self nucleic acids are normally excluded from this compartment (Brencicova and Diebold 2013).
TLR3
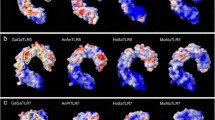
TLR3 recognition of dsRNA (Alexopoulou et al. 2001) recruits the adaptor molecule TRIF to its intracellular TIR domain which leads to the production of interferon and other proinflammatory cytokines. The ectodomain composed of leucine rich repeats (LRR) is involved in ligand recognition. TLR3 detection of RNA agonists has been clarified through crystal structures of human (Bell et al. 2005; Choe et al. 2005) and mouse ectodomains (Liu et al. 2008) in association with dsRNA. The LRR ectodomains form dimers on RNA, and each binds dsRNA at either end of the horseshoe structure (LRR-NT to LRR3 and LRR19 to LRR21), requiring a strand of dsRNA of at least 40–50 nucleotides long (Leonard et al. 2008). Ligand binding brings the ends of two outwardly facing LRRs into close apposition to stabilize the dimer, leading to dimerization of the TIR domains promoting downstream signalling (Liu et al. 2008). TLR3 cleavage by cathepsins in the endosome is required for activation, with a 70-kDa C-terminal fragment (having only one of the ectodomain RNA binding sites) retaining full activity (Garcia-Cattaneo et al. 2012).
TLR signal transduction has been elaborated in mammals and reviewed in detail previously (Kawasaki and Kawai 2014). Upon transduction of a signal through TLR3, the cytoplasmic TIR domain engages TRIF and recruits TRAF6 which ubiquitinates TAK1, phosphorylating MAPKs and transcription factor AP-1, driving transcription of type I IFN. TAK1 also acts on IKK to degrade IκBs, releasing NF-κB. Finally, phosphorylation of TRIF by TBK1 recruits IRF3 which dimerizes and drives transcription of IFN. Most signalling components are present in the chicken genome (Gillespie et al. 2011), and this can be revisited with improved genomic resources. TBK1 is highly conserved in birds, with chTBK1 having 86% identity to human TBK1 (Wang et al. 2017). Knockdown of chTBK1 or duTBK1 using siRNA resulted in reduced expression of IRF7 and IFN-β (Hua et al. 2018; Wang et al. 2017). TRIF has been identified in birds, and chicken and duck TRIF are 67% identical to each other, and only 29–34% identical to the human homologue (Wei et al. 2016; Wheaton et al. 2007). Duck TRIF is upregulated with poly I:C treatment of duck embryonic fibroblasts (Wei et al. 2016) and able to activate IFN-β signalling. Most of the intermediate steps have not been confirmed in birds, so the signalling pathway downstream is inferred from functional data and conserved molecular interactions.
Chicken and Pekin duck TLR3 are 61% and 62% identical to the human ortholog, and share 86% identity to each other (Yilmaz et al. 2005; Zhang et al. 2015). Tissue and cellular expressions of chicken TLRs were systematically examined by RT-PCR (Iqbal et al. 2005a). TLR3 is expressed in most tissues examined including throughout the trachea and intestinal tract and caecum, and many immune tissues including, thymus, spleen, and caecal tonsils, and weakly in bursa. It is expressed in heterophils and highest in CD8 + cells. TLR3 is expressed in chicken embryonic fibroblasts and in immortal cell lines, chicken kidney cells (CKC) and chicken macrophage-like HD-11 cells and weakly in immortalized chicken B cells (DT-40 B cell line). Chicken TLR3 is upregulated by interferon alpha (IFN-α) treatment of leukocytes, and on DF-1 cells, but not chicken HD-11 cells (Karpala et al. 2008). Both mallard and Muscovy ducks have a similar tissue expression profile, which is slightly different from chickens (Jiao et al. 2012). In Pekin ducks, TLR3 is constitutively expressed highest in the trachea, but low in intestine. In Muscovy ducks, TLR3 is constitutively expressed in healthy ducks and basal expression is high in trachea, spleen and pancreas (Jiao et al. 2012).
The function of chicken TLR3 was assessed, as in mammals, using agonists. When heterophils are stimulated with the dsRNA mimic polyinosinic:polycytidylic acid (poly I:C), they show an induced oxidative burst and degranulation (Kogut et al. 2005). Ectopic expression of chicken TLR3 in the human embryonic kidney (HEK293) cell line and stimulation with hTLR3 agonist, poly I:C, activated a reporter assay (Schwarz et al. 2007). Chicken cells respond to poly (I:C) (Karpala et al. 2008), and RNAi-mediated knockdown of TLR3 decreased IFN-β production by DF-1 cells stimulated with IFN-α and poly (I:C) (Karpala et al. 2008). Mallard ducks challenged with poly (I:C) upregulate genes associated with the TLR3 pathway in their transcriptome, and this is confirmed by qPCR analyses (Jax et al. 2021). Chicken DF-1 cells with TLR3, knocked out using CRISPR, could still respond to RNA mimics; however, TLR3/MDA5 double knockout rendered these cells unable to respond to RNA ligands (Lee et al. 2020).
In cell culture, TLR3 induces antiviral effects. Overexpression of chTLR3 in DF-1 cells reduced viral replication and viral titre, implicating TLR3 in a successful response against Newcastle disease virus (NDV) (Cheng et al. 2014). Infection of ducks with two different virulent strains of NDV shows upregulation of TLR3, TLR7 and RLR detectors in the lungs and thymus of NDV-infected ducks (Kang et al. 2015). Activation and upregulation of TLR3 in chicken embryonic fibroblasts (CEFs) through stimulation with poly I:C also inhibited Marek’s disease virus infection (MDV) (Hu et al. 2016). A comparison of several TLR agonists showed 81% inhibition of MDV by poly (I:C), while other agonists gave partial inhibition (Bavananthasivam et al. 2018). Prophylactic TLR agonists can protect chickens from viral infection with several pathogens and, furthermore, are important immunostimulatory adjuvants in vaccine formulations, as reviewed by St Paul et al. (2013).
In other viral infections, TLR3 may be primarily inducing inflammatory responses, sometimes to the detriment of the host. The expression of TLR3 was examined in ducks challenged with duck reovirus, a dsRNA virus that causes necrotic lesions in liver and spleen and 30–40% mortality. Upon infection with duck reovirus, TLR3 is highly induced in the brain, liver and spleen, but not in the lung until 3 dpi (Zhang et al. 2015). In the duck fibroblast cell line CCL141, the upregulation of TLR3 correlated with viral load. Chickens infected with an H5N1 highly pathogenic avian influenza strain showed upregulation of TLR3 in the lung (Ranaware et al. 2016) or lung and brain (Karpala et al. 2008). Chickens infected with a highly pathogenic H7N1 strain had increased TLR3 expression in the lung, the spleen and especially the brain (Cornelissen et al. 2013). Unfortunately, duck TLR3 was not included in the direct comparisons in that study, which would have been interesting because ducks typically suffer less pathology. In Pekin ducks, TLR3 is upregulated in all tissues tested following highly pathogenic H5N1 infection in the lung, spleen and intestine (Campbell et al. 2021). Muscovy ducks infected with an H5N1 strain showed upregulation of TLR3 in the brain, but not in the spleen or lungs (Jiao et al. 2012). Similarly, infection of Muscovy ducks with two other H5N1 strains showed upregulation in brain, with downregulation in the lungs (Wei et al. 2013). Muscovy ducks are more susceptible to H5N1 virus infection (Pantin-Jackwood et al. 2013). Notably, in mice infected with an H3N2 strain of influenza, TLR3 was highly upregulated in the lungs and mice showed increased inflammation in comparison with TLR3 knockout mice, and knockout mice survived longer (Le Goffic et al. 2006).
TLR7
TLR7 recognizes ssRNA (Lund et al. 2004) and small chemical compounds mimicking nucleosides. Mammalian TLR7 can be stimulated by loxoribine and imiquimod, while resiquimod (R848) can stimulate both TLR7 and TLR8 (Hemmi et al. 2002; Jurk et al. 2002). Murine TLR7 is present on plasmacytoid dendritic cells (pDCs), also called professional interferon producing cells (IPCs) (Liu 2005), and is involved in the detection of single-stranded RNA viruses like influenza (Diebold et al. 2004). Detection of influenza virus requires the intact hemagglutinin receptor indicating that entry occurs through receptor mediated endocytosis and leads to the production of IFN-α. Interferon production is dependent on TLR7 and MyD88 and can be blocked by chloroquine, which stops endosomal trafficking (Diebold et al. 2004). TLR7 dimerizes and must be cleaved within the extracellular domain in the endosome for generation of the downstream signal, but the parts remain associated through a disulphide bond and are both involved in ligand discrimination (Kanno et al. 2013). Mammalian TLR7 recognition involves two sites: one for small agonists (or guanosine) and the second for uridine containing ssRNA (Zhang et al. 2016). TLR7 exists as a monomer in the absence of ligands, but dimerization is induced by R848 alone or by the combination of guanosine and poly U RNA. Ligand binding at the first site bridges the two TLRs involving LRR8, and LRR11-14 of one TLR7, and LRR16*-18* of the second and the same for the second TLR7 molecule (Zhang et al. 2016). Disulphide bonds create the secondary structure and residues of a Z-loop, and LRR1-5 and LRR20* generate the surface for binding ssRNA (Zhang et al. 2016). Further crystallographic studies identified guanosine 2′3′-cyclic phosphate (2′3′-cGMP) as a possible endogenous ligand for site 1 and demonstrate affinity for uridine containing RNAs at site 2 is greater if there are successive uridines (Zhang et al. 2018b).
Chicken and duck TLR7 are 62% and 66% identical to human TLR7 and 85% identical to each other (MacDonald et al. 2008; Philbin et al. 2005). Chicken splenocytes respond to TLR7/8 agonists (including R848 and loxoribine, poly U RNA and ssRNAs) with the production of proinflammatory cytokines, but not interferons (Philbin et al. 2005). The chicken macrophage–like cell line HD-11 also responded with the production of IL-1β, but not interferons. This appears to be strain dependent in chickens, with some lines producing interferon in response to loxoribine (Kogut et al. 2006). Duck splenocytes stimulated with imiquimod produce both proinflammatory cytokines and IFN-α. Confirming chTLR7 is also endosomally located; treatment of chicken splenocytes with chloroquine completely abrogates the production of IL-1β, in response to R848 (Philbin et al. 2005).
Signalling downstream of TLR7 involves MyD88 and TRAF6 recruitment to the myddosome described above (Fitzgerald and Kagan 2020). Duck MyD88 showed 88% similarity to chicken MyD88 (Cheng et al. 2015b). Overexpression induced activation of NF-κB and IL-6. Duck and chicken TRAF6 share 96.5% amino acid identity (** et al. 2017)(Zhai et al. 2015). Overexpression of duck TRAF6 activated NF-κB, and knockdown impaired poly (I:C) or SeV stimulated activation of NF-kB (Zhai et al. 2015). The duck IKKα and IkB subunits of IkB kinase (IKK) were cloned, and overexpression of either initiated gene expression of IFN-β, while knockdown inhibited Sendai virus induced NF-kB activation (Li et al. 2021; Zhou et al. 2021).
TLR7 and 8 are adjacent in most vertebrates, clearly the products of an ancient duplication. TLR8 is disrupted in the genome of both chickens (Philbin et al. 2005) and ducks (MacDonald et al. 2008). Indeed, it appears that intact TLR8 is not found in avian genomes (Velova et al. 2018). Some birds, however, show duplication of TLR7 genes (Velova et al. 2018). Duplicated TLR7s appear in 8 different species of birds, which are in different phylogenetic clades. Duplicated TLR7 genes appear in killdeer, cuckoo and passerines including crows and zebra finch (Velova et al. 2018). In each case, these sequences differ from one another by as much as 21 amino acids, but these sequences only group with the gene from the same species. They also do not show any evidence of gene conversion. The authors conclude that TLR7 has been duplicated many times and independently in each species (Velova et al. 2018). Ruddy turnstones may have three copies of TLR7 since five allelic sequences were identified in some individuals (Raven et al. 2017). Birds do not have an equivalent of the mammalian TLR9, which is phylogenetically close to TLR7 and TLR8 (Roach et al. 2005).
Chicken TLR7 is expressed in the spleen, bursa, caecal tonsil and most intestinal tissues (Iqbal et al. 2005a). It is strongly expressed in primary B cells and other leukocytes (Philbin et al. 2005) and expressed in the HD11 and DT40 immortalized cell lines (Philbin et al. 2005). Duck TLR7 is highly expressed in the spleen, bursa and lung (MacDonald et al. 2008). High expression in the lung was not seen in chicken.
Mammalian TLR7 is expressed in plasmacytoid dendritic cells (pDCs) and is involved in the production of IFN-α by these professional interferon producing cells (Diebold et al. 2004). Intriguingly, small round intraepithelial haematopoetic cells in duck ileum have the hallmarks of pDCs, as they are TLR7-positive cells, detected by in situ hybridization, and expand in numbers in response to R848 agonist or infection with avian influenza (Volmer et al. 2011). These cells are the same shape and in the same location as IFN-α-positive cells, suggesting the cells are double positive for TLR7 and IFN-α. These data suggest that potentially pDCs as professional interferon–producing cells are conserved in birds. Other types of dendritic cells have been characterized in birds (Nagy et al. 2016; Wu et al. 2010), but not pDCs.
Viral antagonism of TLR signalling
Many bacterial and viral proteins antagonize human TLR signaling at almost every step (Rosadini and Kagan 2015). A few examples of RNA viruses blocking avian TLR signalling are beginning to emerge. Comparison of three different Newcastle disease virus (NDV) strains showed that the most virulent NDV strain decreases TLR7 expression in HD11 cells, although IFN-α and IFN-β increased for this strain (Zhang et al. 2018a). Prior treatment of cells with loxoribine, a TLR7 agonist, decreases viral replication. Infectious bronchitis virus (IBV) inhibits activation of the TLR7 pathway by downregulating transcription, but upregulates TLR3 (Zhu et al. 2020). Activation of TLR7 with agonists decreased IBV replication in chicken embryo kidney cells, but inhibition of TLR3 increased viral titres.
Evolution of avian TLR3 and 7
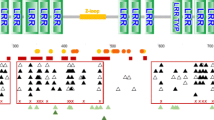
A comprehensive study of all avian TLR receptors in which sequences were available (Velova et al. 2018) showed low levels of positive selection acting on the endosomal receptors compared to others, presumably because of the conserved nature of the nucleic acid ligands. Across diverse species, TLR3 showed a number of conservative positive selected sites across the three protein domains, with most in the extracellular domain, including 3 that were non-conservative differences, changing the physiochemical nature of that residue. In TLR7, there were more PSS with most being conservative; however, there were 7 sites of non-conservative changes. Five of these were located near the cleavage site, where TLR7 is cleaved for signal transduction (Kanno et al. 2013). Two sites in TLR3 were near the dimerization site, and since dimerization depends on RNA length, selection on these residues may influence binding. Many of the sites identified in these receptors will be important to consider in future analyses of bird populations and pathogen selection.
A comparison of TLR sequences from the galloanseriform clade looked at interspecific differences within TLR4, TLR5 and TLR7 sequences (Vinkler et al. 2014). There were stronger signatures of positive selection acting on the ligand binding region of TLR4 and TLR5, compared to TLR7. This likely reflects that TLR4 and TLR5 are recognizing structurally variable bacterial ligands, while TLR7 is recognizing a conserved molecular structure. In TLR7, 4 positively selected sites were identified, and 25 non-conservative sites were shown to be diversifying between species. Some of the non-conservative sites are potentially important; however, for TLR7, most are in the part of the extracellular domain that is cleaved in the priming step for signalling, one in the signal peptide and one in the TIR domain. A systematic analysis of 66 representative bird species, covering 26 orders, identified 9 positively selected sites in TLR3, and 25 in TLR7 (Yang et al. 2021). Several of these sites were identified using two different prediction methods.
In a study aimed at identifying differences in RNA detectors in shorebirds, Raven and colleagues amplified a fragment of TLR3 and TLR7 from birds from three species, the sanderling (Calidris alba), red-necked stint (C. ruficollis) and ruddy turnstone (Arenaria interpres) (Raven et al. 2017). They included sequences from 50 available bird genomes. TLR3 and TLR7 showed overall a predominance of purifying selection. However, there were a small number of positively selected sites in TLR3 (3.2%) and TLR7 (2.5%). Sites under positive selection appear to be in the LRR domains. In the population data, for TLR3, they found six sites were under positive selection, with half of these being in LRR14 in one species (C. alba). In TLR7, there were 14 sites under positive selection, and several of these were shared in more than one species (Raven et al. 2017). Interestingly, even domestic chicken breeds show substantial allelic diversity of TLR genes (Świderská et al. 2018). For example, there were 18 non-synonymous single nucleotide variants or alleles of TLR3 and 7 alleles of TL7. There was also evidence of convergent evolution leading to the same allele of TLR7 in unrelated breeds. In all, the level of avian TLR polymorphism far exceeds the polymorphism found in human TLRs.
RLR receptors and activation through MAVS oligomerization
A family of pattern recognition receptors named for the prototype retinoic acid gene-I (RIG-I) is called the RIG-I-like receptor (RLR). Two other members of this family include melanoma differentiation–associated factor 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2). The RLR receptors detect viral RNA in the cytoplasm to initiate a signalling cascade leading to type I interferon, turning on hundreds of interferon stimulated genes (ISGs). RIG-I-like receptors are involved in detection of a number of RNA viruses (Loo and Gale 2011; Rehwinkel and Gack 2020). RIG-I has specificity for nascent viral RNA that forms panhandle structures with a 5′-triphosphate overhang (Hornung et al. 2006; Liu et al. 2015a; Schlee et al. 2009a, 2009b). In an influenza virus infection, these RIG-I agonists are generated as short minigenome viral RNAs as a result of RNA polymerase dysregulation during replication (Te Velthuis et al. 2018). In mouse and man, RIG-I and MDA5 detect distinct viruses (Kato et al. 2006). RIG-I is involved in detection of short ds RNA and single-stranded RNA viruses with panhandle structures like Sendai virus, influenza virus, hepatitis C, ebolavirus and others. MDA5 has specificity for long double-stranded RNA and is involved in the detection of picornaviruses and vaccinia virus. Both receptors are required for detection of other viruses like West Nile virus. RIG-I and MDA5 selectively recognize short and long dsRNA of synthetic or viral origin depending on length, respectively (Kato et al. 2008). MDA5 measures RNA length in the range of 0.5 to 7 kb by forming and disassembling filaments depending on ATP hydrolysis (Peisley et al. 2012). Sun Hur has critically reviewed the details of recognition by RLRs and how this prevents the discrimination between host and viral RNA (Hur 2019). The RLR receptor LGP2 has no CARD domains, and thus cannot engage mitochondrial antiviral signalling (MAVS), and has been implicated in a confusing array of positive or negative regulatory roles, reviewed recently (Rehwinkel and Gack 2020). This has been clarified recently as its role as an inhibitor of DICER has come to light (van der Veen et al. 2018). DICER is the mediator of RNA interference pathways of destruction of double-stranded RNA, an ancient antiviral pathway which is not active in cells with a functional interferon response (Maillard et al. 2019). Since many RNA viruses also have avian hosts, this family of receptors are critically involved in detecting viruses relevant to birds such as influenza A viruses, NDV and WNV.
RIG-I
A RIG-I receptor was identified in ducks and shares only 53% identity to human RIG-I (Barber et al. 2010). However, it conserves the structure of two N-terminal caspase activation and recruitment domains (CARD), a central helicase domain and a C-terminal regulatory domain. The C terminal domain recognizes the ligand, while the N-terminal tandem CARD domains recruit the MAVS protein. Remarkably, a RIG-I homologue could not be identified in the chicken genome sequence derived from the red junglefowl. It was also not found among the available transcriptome sequences, while the same approach was used to identify the zebra finch RIG-I sequence. The duck RIG-I sequence could functionally compensate to allow recognition of RIG-I ligand in chicken cells (Barber et al. 2010). Subsequent examination of 62 bird genomes for RLR sequences also failed to identify RIG-I in chicken, Japanese quail or turkey (Zheng and Satta 2018). Together, these data support the idea that RIG-I has been lost in the galliform bird lineage, or it has undergone significant sequence divergence. The region has retained some synteny as the flanking aconitase 1 gene can be found; however, the gene order and organization differ between ducks and chickens. The apparent lack of any gene in birds must be considered with the caveat that avian genomes are not completely assembled, typically were built using the chicken genome as scaffold, and high GC content regions are notoriously difficult to sequence by high throughput sequencing (Hron et al. 2015).
The duck RIG-I was crystallized before other RIG-I sequences (Kowalinski et al. 2011), which can probably be attributed to unique features of the sequence. Notably, duck RIG-I lacks the critical lysine residue K172, the site where TRIM25 covalently attaches polyubiquitin chains for activation of human RIG-I CARD domains (Gack et al. 2007). Mutation of potential ubiquitin sites in the duck CARD domains showed ubiquitin is attached at K167 and K193, but the double mutant is still active (Miranzo-Navarro and Magor 2014). Ubiquitin chains stabilize the structure of the CARD domains for ligation with MAVS. Duck RIG-I, like mammalian RIG-I (Zeng et al. 2010), can associate with unanchored ubiquitin chains allowing the formation of the crystal structure. Availability of the duck RIG-I crystal structure enabled prediction and mutation of critical residues involved in the interaction with MAVS, and we performed experiments alongside mutational analyses of human RIG-I, allowing us to contribute to the elucidation of a general model of activation of RIG-I (Wu et al. 2014). The RIG-I CARD domains from four bound molecules form a tetrameric lock-washer type structure offset by one CARD domain, upon which the single CARD domain of MAVS can bind and begin the process of oligomerization. Then, MAVS oligomerizes and assembles into a helical filament which initiates signal transmission. Mutants that cannot oligomerize are unable to activate the interferon reporter; thus, oligomerized MAVS is required to recruit TRAF6. This mechanism is highly conserved despite weak sequence conservation of duck and human MAVS, having only 28% sequence identity. More recently, RIPLET was shown to be more important in the activation of RIG-I in mammalian cells (Hayman et al. 2019), and it binds to RIG-I once it has bound to dsRNA, allowing discrimination depending on RNA length (Cadena et al. 2019). However, RIPLET appears to be missing in chickens, and ducks primarily express an incomplete protein lacking the RING domain (Magor et al. 2013b), although a full-length version is present in the genome. An additional TRIM protein, TRIM27-L, is present in ducks but not chickens or turkeys, and co-expression with the constitutively active duck RIG-I N-terminal end (CARD domains) greatly augments activation of the MAVS signalling pathway in chicken DF-1 cells (Blaine et al. 2015). Activation of human MDA5 requires the E3 ligase TRIM65 (Kato et al. 2021; Lang et al. 2017). Comparison of mammalian RLR-TRIM contact sites determined by crystallography suggest that well-conserved sites in RIPLET/TRIM65 contact well-conserved sites in RIG-I/MDA5, respectively, while less conserved sites interact with less conserved sites, suggesting that these pairs of proteins have co-evolutionary relationships (Kato et al. 2021). The mechanism of activation of avian RIG-I and MDA5 remains unresolved.
RIG-I involvement in antiviral responses in birds is inferred from upregulation of RIG-I during infection with highly pathogenic avian influenza in ducks (Barber et al. 2010) and NDV infection of geese (Sun et al. 2013b). Microarrays from chicken cells, transfected with duck RIG-I and infected with influenza viruses, show augmented expression of many of ISGs in the cells with duck RIG-I in comparison to vector only (Barber et al. 2013). Infection of ducks, chickens and quail with highly pathogenic H5N1 were carried out, and transcriptomes compared to reveal a cluster of 189 genes upregulated in ducks, but not quail or chickens (Morris et al. 2020). These genes are strongly enriched for the RIG-I pathway and ISGs. Our recent transcriptome analyses of ducks infected with high and low pathogenic influenza mirror these results with RIG-I pathway genes being important in both the lung and the intestine, sites of infection (Campbell et al. 2021). Among the top 100 genes in the lung following infection with H5N1 are type I interferons and ISGs.
Duck RIG-I transfected into chicken DF-1 cells can initiate signalling and induce antiviral proteins including IFN-β, Mx and PKR (Barber et al. 2010). Similarly, pigeon and goose RIG-I are functional in DF-1 cells (Xu et al. 2015). Goose RIG-I lacks a serine at residue 8 which is phosphorylated for autorepression in mouse and man (Nistal-Villan et al. 2010). The mutant G8S, restoring this site, induces much less interferon than wild-type goose RIG-I (Xu et al. 2015). This residue is not well conserved in birds.
Independent of stimulating interferon pathways, mammalian RIG-I can directly restrict incoming influenza virus particles (Weber et al. 2015). Binding of RIG-I to the incoming influenza ribonucleoprotein (RNP) is altered by the residue 627 in PB2. RNPs with PB2 627 K in mammalian strains are bound inefficiently, while RNPs with the avian glutamate are efficiently bound by RIG-I. This function does not require signalling capacity of RIG-I, and was lost with deletion of RIG-I but not MAVS. Chickens, lacking RIG-I, may allow greater viral replication by the absence of this direct restriction of incoming virus.
TRIM25 can also be a viral restriction factor. Mammalian TRIM25 has capacity to directly restrict influenza virus in the nucleus by binding to viral RNPs and prevents the viral RNA from moving into the polymerase complex (Meyerson et al. 2017). This activity is independent of the ubiquitin ligase activity or the interferon pathway. Duck TRIM25 is 69% similar to chicken TRIM25 and only 48% identical to the human ortholog. Overexpression of TRIM25 in chicken cells does inhibit replication of avian leukosis virus, and siRNA knockdown increases viral titre (Zhou et al. 2020). It is not known whether this is due to direct antiviral action or TRIM25 acting on chMDA5, but TRIM25 is not known to activate mammalian MDA5. Similarly, knockdown of TRIM25 in duck cells increases the viral titre of DTMUV (Kaikai et al. 2021). TRIM25 is a direct restriction factor for infectious bursal disease virus (IBDV), which ubiquitinates viral protein 3 (VP3) marking it for degradation (Wang et al. 2021).
MDA5
MDA5 is a detector for long double-stranded RNA and synthetic RNAs like poly (I:C). MDA5 detects RNA involving both its helicase domain and the C terminal domain, and clustering of CARD domains again allows engagement of MAVS. The length of the RNA ligand is essential, as MDA5 stacks along the length of the RNA and the tandem CARD domains oligomerize and recruit MAVS (Peisley et al. 2012; Wu et al. 2013). Again, the oligomerization step is essential for activation of MAVS and recruitment of TRAF6 to initiate the downstream signaling cascade.
Chicken MDA5 shares around 60% identity with human MDA5 (Karpala et al. 2011), with most of the difference concentrated in the CARD domain (43% identity)(Childs et al. 2007). Chicken MDA5 can activate an interferon inducible reporter in chicken DF-1 cells (Childs et al. 2007). MDA5 is upregulated by treatment with type I interferon or influenza infection suggesting it is involved in detecting this virus, but knockdown does not alter viral replication kinetics (Karpala et al. 2011). However, chMDA5 does affect the response to the virus, since silencing MDA5 decreases IFN-β production in infected cells (Liniger et al. 2012). This suggests that MDA5 does functionally compensate for the loss of RIG-I in chickens. Chicken MDA5, distinct from mammalian MDA5, preferentially responds to short dsRNA (Hayashi et al. 2014), although it can also respond to long poly (I:C) (Barber et al. 2010; Karpala et al. 2011). Indeed, it can recognize in vitro transcribed RNA with or without 5′-triphosphate at the ends (Karpala et al. 2011). Chicken MDA5 can also recognize NDV, showing sites of positive selection in the DECH helicase domain and also in LGP2 (Xu et al. 2019). One positive selected site L625 was swapped back to the consensus avian glutamate, L625E, and this mutant had lower affinity for NDV RNA. LGP2 augmented this interaction. Three additional positive selected sites in LGP2 were similarly shown to increase interaction with NDV RNA (Xu et al. 2019). Duck MDA5 does not share these residues (Barber et al. 2010) and is unlikely to bind NDV RNA. Duck MDA5 is upregulated in influenza infected tissues (Fleming-Canepa et al. 2019), and over-expression of the CARD domain can decrease viral titres (Wei et al. 2014).
Conservation of MAVS in birds
MAVS is critically involved in the production of NFκB and IRF3 in multiple cell types, except pDCs, as knockout mice failed to respond to poly (I:C) and were severely compromised in immune defense against viruses (Sun et al. 2006). Knockdown of MAVS in ducks significantly reduced induction of IFN-β following poly (I:C) or Sendai virus infection (Li et al. 2016). MAVS sequences are very poorly conserved throughout vertebrates. Duck and chicken MAVS are 58% identical, and duck and human MAVS are only 28% identical (**ao et al. 2020). Of the many potential post-translational modification sites in human MAVS, only a few are conserved in birds. These include the lysines K7 and K442 (numbering based on human sequence) where ubiquitination to target MAVS for degradation occurs. Sequences within the CARD domain are conserved between avian and human MAVS (Sun et al. 2019), a T54 and GWV at residues 67–69, sites involved in activation of human MAVS (Seth et al. 2005). The duck and chicken MAVS conserve sequences around a putative pLxIS motif (**ao et al. 2020). This motif, which is DLAIS in human MAVS, is phosphorylated by IKKβ, which in turn recruits TBK1 that phosphorylates IRF3 for activation. Upon activation, IRF3 undergoes dimerization and entry into the nucleus where it is involved in the production of type 1 interferon. Even the sequence of this site is not well conserved in birds and hints at the selective pressures of pathogens on this protein.
Viral antagonism of RLR receptors
In mammals, there are many viral proteins that directly interfere with signaling through MAVS, either by cleaving it or modulating function as a strategy to overcome host responses, as recently reviewed (Ren et al. 2020). While very few studies look at this in birds, as these pathways are studied, examples emerge. In fact, chicken MDA5 was characterized initially as a target for interference by avian and human paramyxoviruses, through the V protein (Childs et al. 2007). Influenza NS1 shuts down interferon signalling through interaction with TRIM25 and RIPLETs, the E3 ubiquitin ligases that activate RIG-I. This has been shown to be strain and species dependent. Influenza NS1 protein from four different strains inhibit human TRIM25, but only one interacts with chicken TRIM25 (Rajsbaum et al. 2012). Another influenza protein derived from the second reading frame of polymerase B1 called PB1-F2 inhibits MAVS signalling induced by overexpressed duck MAVS or RIG-I in chicken DF-1 cells (**ao et al. 2020). We showed an interaction between duck MAVS and PB1-F2 through co-immunoprecipitation from these cells. This interaction also blocks the TRIM25 mediated RIG-I CARD ubiquitination, possibly through loss of mitochondrial membrane potential. The NS1 protein of the flavivirus duck Tembusu virus (DTMUV) also blocks MAVS signalling by interacting with the C-terminal region of MAVS (Wu et al. 2019). This effectively sequesters MAVS from signalling downstream of RIG-I or MDA5 in the duck cells, accounting for the weak induction of interferon by infection with DTMUV.
Evolution of avian RLRs
Analysis of the evolution of RIG-like receptors in 62 avian species highlights the conserved and evolving features of the three RLRs (Zheng and Satta 2018). The conservation scores and dN/dS ratios indicate that avian MDA5 is highly conserved in the helicase domain, with lower scores in the CARD domains. MDA5 shows several sites of positive selection in the CARD domain, while RIG-I shows only one potential site. Two sites showing positive selection at either ends of the helicase domain are located at identical places in both RIG-I and MDA5, although the functional significance of this variation is not known. LGP2 has the lowest conservation score of the three RLR receptors but shows unique sites of positive selection only in the three species for which RIG-I was not found (chicken, turkey and quail). With the RLRs, almost all the positively selected sites are exposed, and none is located where they would alter contact with the RNA. Most likely, these sites may affect regulation of the receptor and not detection of RNA ligands. Functionally important sites were analysed, including the two ubiquitination sites identified in ducks (K167 and K193). Of these K167 is invariant in all sequences, while K193 has substitutions to threonine in two crow species and the carmine bee-eater. Finally, the only positively selected site in RIG-I is located the region that can be spliced out in a splice variant that is created in duck (Miranzo-Navarro and Magor 2014), and humans (Gack et al. 2008), which renders RIG-I inactive.
STING
Stimulator of interferon gene (STING) is an adaptor protein critically involved in the detection of DNA and RNA viruses and initiation of NF-κB signalling and IRF3 transcription (Ishikawa and Barber 2008). STING is the key adaptor involved in the cyclic GMP-AMP synthase (cGAS) signalling pathway, primarily a cytoplasmic DNA sensor (Sun et al. 2013a). Upon detection of cGAMP, STING triggers phosphorylation of IRF3 and TBK1. Several experiments showed that STING is also involved in defense against RNA viruses (Holm et al. 2016; Schoggins et al. 2014). cGAS knockout mice were more susceptible to West Nile virus, and infected cGAS−/− bone marrow macrophages had higher viral load than wild-type and lower expression of IFNβ and all ISGs (Schoggins et al. 2014). In mouse embryonic fibroblasts, lack of STING (but not cGAS) significantly reduced the IFN produced upon infection with NDV and Sendai virus (SeV) (Holm et al. 2016). Co-immunoprecipitation experiments demonstrated that STING interacts with RIG-I and MAVS, but not MDA5 (Ishikawa and Barber 2008). Infection with SeV greatly increased the interaction between STING and RIG-I. These studies suggest a role for STING signalling, perhaps involving endogenous ligands, in protection against RNA viruses.
Because chickens lack RIG-I, it was of interest whether chicken STING is involved in RNA virus detection in chickens. Indeed, chicken STING is capable of activation of the IFN pathway in chicken DF-1 cells (Cheng et al. 2015a). Overexpression of STING in DF-1 cells activates IRF-7 signalling and NF-κB and an IFN-β reporter. STING is upregulated in these cells upon infection with NDV, AIV or fowlpox virus FPV, and upon knockdown of STING, the titre of all three viruses increased. Duck STING has recently been characterized, and overexpression in duck embryonic fibroblasts increases IFN-β promoter activity (Cheng et al. 2019a). Constitutive expression of duck STING is high in stomach and intestinal tissues and also in the trachea and lung (all relevant in influenza virus infections). It is also upregulated in duck embryonic fibroblasts and in tissues of ducks infected with a low pathogenic H9N2 influenza strain. This suggests that STING may be involved in viral defense in ducks. It is not yet known whether STING interacts with duck RIG-I or MDA5.
Chicken and duck STING share only 43% identity to human STING and 71% amino acid sequence identity to each other (Cheng et al. 2019a, 2015a). The overall structure of STING is conserved, having four predicted transmembrane domains, two c-di-GMP binding domains and a carboxy-terminal tail (Cheng et al. 2019a). The location of the pLxIS motif in the C-terminal tail can be predicted, although the sequence shows considerable divergence compared to the human sequence ELLIS, as SLQVS in chickens (Cheng et al. 2019b). The site SLQIS was functionally characterized in duck STING (Zhang et al. 2020). Phosphorylation at this serine is required for recruiting TBK1 and signalling through IRF3. Because birds lack IRF3 (Magor et al. 2013b), signalling was presumed to involve IRF7. By making an IRF7 knockout in chicken DF-1 cells, Cheng and colleagues showed that IRF7 is indispensable for signalling through MAVS and STING in chicken cells (Cheng et al. 2019b).
RNA virus antagonism of avian STING
Numerous viral proteins are capable of inhibiting the STING pathway as reviewed recently (Eaglesham and Kranzusch 2020; Ma and Damania 2016). While this has been relatively unexplored in avian cells, some examples have been noted. Several RNA viruses affecting human STING, also circulate in birds, notably influenza A viruses. The influenza A virus hemagglutinin fusion peptide can directly inhibit human STING (Holm et al. 2016). Fusion peptide inhibits STING activation triggered by membrane fusion, a function of STING previously noted (Holm et al. 2012). It is likely that detection of damage by viral entry into membranes is a fundamental function of evolutionarily ancient STING. The flavivirus DTMUV cleaves STING using protease NS2B3, contributing to this virus’ arsenal of ways to inhibit IFN-β transcription (Wu et al. 2019). Finally, DTMUV strongly blocks the signalling pathway by binding STING with non-structural protein NS2A, interfering with the binding of TBK1 (Zhang et al. 2020). A functional screen of the oncogenic herpesvirus that causes Marek’s disease (a DNA virus) identified five proteins that can inhibit the STING pathway, including one called Meq, that blocks recruitment of TBK1 to STING in chicken cells (Li et al. 2019). These examples demonstrate the vulnerability of this point in the signalling pathways.
Conclusions
Great strides have been made in establishing the function of avian PRR involved in detection and signalling in response to RNA viruses. Several studies have identified sites within these receptors that are under positive selection, and some are non-conservative changes. These sites will be important to consider in the context of populations and pathogens. It will also be interesting to consider evolution of the PRR receptors and their adaptors TRIF, MAVS and STING between birds of different species, which may emerge from the avian genome projects. It is largely unknown whether RLR receptors and adaptors show variation within bird populations contributing to pathogen susceptibility. Considerable effort is needed to understand the regulation of these signalling pathways in any species, including birds. Finally, much remains to be done to understand viral antagonism of these pathways in birds. The number of ways that viruses interfere in these pathways is undoubtedly as great in birds as it is in mouse and man.
References
Afonso CL (2021) Virulence during Newcastle disease viruses cross species adaptation. Viruses 13:e110
Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-kappaB by toll-like receptor 3. Nature 413:732–738
Barber MR, Aldridge JR Jr, Fleming-Canepa X, Wang YD, Webster RG, Magor KE (2013) Identification of avian RIG-I responsive genes during influenza infection. Mol Immunol 54:89–97
Barber MRW, Aldridge JR Jr, Webster RG, Magor KE (2010) Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A 107:5913–5918
Bavananthasivam J, Kulkarni RR, Read L, Sharif S (2018) Reduction of Marek’s disease virus infection by toll-like receptor ligands in chicken embryo fibroblast cells. Viral Immunol 31:389–396
Bell JK, Botos I, Hall PR, Askins J, Shiloach J, Segal DM, Davies DR (2005) The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc Natl Acad Sci U S A 102:10976–10980
Blaine AH, Miranzo-Navarro D, Campbell LK, Aldridge JR Jr, Webster RG, Magor KE (2015) Duck TRIM27-L enhances MAVS signaling and is absent in chickens and turkeys. Mol Immunol 67:607–615
Boyd A, Philbin VJ, Smith AL (2007) Conserved and distinct aspects of the avian Toll-like receptor (TLR) system: implications for transmission and control of bird-borne zoonoses. Biochem Soc Trans 35:1504–1507
Boyd AC, Peroval MY, Hammond JA, Prickett MD, Young JR, Smith AL (2012) TLR15 is unique to avian and reptilian lineages and recognizes a yeast-derived agonist. J Immunol 189:4930–4938
Brencicova E, Diebold SS (2013) Nucleic acids and endosomal pattern recognition: how to tell friend from foe? Front Cell Infect Microbiol 3:37
Brownlie R, Allan B (2011) Avian toll-like receptors. Cell Tissue Res 343:121–130
Brownlie R, Zhu J, Allan B, Mutwiri GK, Babiuk LA, Potter A, Griebel P (2009) Chicken TLR21 acts as a functional homologue to mammalian TLR9 in the recognition of CpG oligodeoxynucleotides. Mol Immunol 46:3163–3170
Cadena C, Ahmad S, Xavier A, Willemsen J, Park S, Park JW, Oh SW, Fujita T, Hou F, Binder M, Hur S (2019) Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 177:1187-1200.e16
Campbell LK, Magor KE (2020) Pattern recognition receptor signaling and innate responses to influenza A viruses in the mallard duck, compared to humans and chickens. Front Cell Infect Microbiol 10:209
Campbell LK, Fleming-Canepa X, Webster RG, Magor KE (2021) Tissue Specific Transcriptome Changes Upon Influenza A Virus Replication in the Duck. Frontiers in Immunology 12 https://doi.org/10.3389/fimmu.2021.786205
Chen S, Cheng A, Wang M (2013) Innate sensing of viruses by pattern recognition receptors in birds. Vet Res 44:82
Chen S, Wang T, Liu P, Yang C, Wang M, Jia R, Zhu D, Liu M, Yang Q, Wu Y, Zhao X, Cheng A (2019) Duck interferon regulatory factor 7 (IRF7) can control duck Tembusu virus (DTMUV) infection by triggering type I interferon production and its signal transduction pathway. Cytokine 113:31–38
Cheng J, Sun Y, Zhang X, Zhang F, Zhang S, Yu S, Qiu X, Tan L, Song C, Gao S, Wu Y, Ding C (2014) Toll-like receptor 3 inhibits Newcastle disease virus replication through activation of pro-inflammatory cytokines and the type-1 interferon pathway. Arch Virol 159:2937–2948
Cheng Y, Liu Y, Shi S, Niu Q, Zhu W, Wang Z, Ma J, Wang H, Yan Y, Sun J (2019a) Functional Characterization of Duck STING in IFN-β Induction and Anti-H9N2 Avian Influenza Viruses Infections. Front Immunol 10:2224
Cheng Y, Sun Y, Wang H, Yan Y, Ding C, Sun J (2015a) Chicken STING mediates activation of the IFN gene independently of the RIG-I gene. J Immunol 195:3922–3936
Cheng Y, Wang H, Yan Y, Ding C, Sun J (2015b) Two myeloid differentiation factor 88 (MyD88) isoforms identified in ducks. Dev Comp Immunol 52:144–154
Cheng Y, Zhu W, Ding C, Niu Q, Wang H, Yan Y, Sun J (2019b) IRF7 Is involved in both STING and MAVS mediating IFN-β signaling in IRF3-involved in both. J Immunol 203:1930–1942
Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S (2007) mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200
Choe J, Kelker MS, Wilson IA (2005) Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309:581–585
Chu DK, Leung CY, Gilbert M, Joyner PH, Ng EM, Tse TM, Guan Y, Peiris JS, Poon LL (2011) Avian coronavirus in wild aquatic birds. J Virol 85:12815–12820
Cormican P, Lloyd AT, Downing T, Connell SJ, Bradley D, O’Farrelly C (2009) The avian Toll-Like receptor pathway–subtle differences amidst general conformity. Dev Comp Immunol 33:967–973
Cornelissen JBWJ, Vervelde L, Post J, Rebel JMJ (2013) Differences in highly pathogenic avian influenza viral pathogenesis and associated early inflammatory response in chickens and ducks. Avian Pathology: Journal of the W.V.P.A 42:347–364
de Zoete MR, Bouwman LI, Keestra AM, van Putten JP (2011) Cleavage and activation of a Toll-like receptor by microbial proteases. Proc Natl Acad Sci U S A 108:4968–4973
Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C (2004) Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529–1531
Eaglesham JB, Kranzusch PJ (2020) Conserved strategies for pathogen evasion of cGAS-STING immunity. Curr Opin Immunol 66:27–34
Evseev D, Magor KE (2019) Innate immune responses to avian influenza viruses in ducks and chickens. Vet Sci 6:e5
Fitzgerald KA, Kagan JC (2020) Toll-like receptors and the control of immunity. Cell 180:1044–1066
Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496
Fleming-Canepa X, Aldridge JR Jr, Canniff L, Kobewka M, Jax E, Webster RG, Magor KE (2019) Duck innate immune responses to high and low pathogenicity H5 avian influenza viruses. Vet Microbiol 228:101–111
Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU (2008) Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci USA 105:16743–16748
Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920
Garcia-Cattaneo A, Gobert FX, Müller M, Toscano F, Flores M, Lescure A, Del Nery E, Benaroch P (2012) Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc Natl Acad Sci U S A 109:9053–9058
Gillespie M, Shamovsky V, D’Eustachio P (2011) Human and chicken TLR pathways: manual curation and computer-based orthology analysis. Mamm Genome 22:130–138
Grant CE, Vasa MZ, Deeley RG (1995) cIRF-3, a new member of the interferon regulatory factor (IRF) family that is rapidly and transiently induced by dsRNA. Nucleic Acids Res 23:2137–2146
Guo SY, Li C, Dai XM, Zhao C, Chang WS (2014) Distribution of the type I interferon in different organs of chicken digestive system. Cent Eur J Immunol 39:455–460
Hayashi T, Watanabe C, Suzuki Y, Tanikawa T, Uchida Y, Saito T (2014) Chicken MDA5 senses short double-stranded RNA with implications for antiviral response against avian influenza viruses in chicken. J Innate Immun 6:58–71
Hayman TJ, Hsu AC, Kolesnik TB, Dagley LF, Willemsen J, Tate MD, Baker PJ, Kershaw NJ, Kedzierski L, Webb AI, Wark PA, Kedzierska K, Masters SL, Belz GT, Binder M, Hansbro PM, Nicola NA, Nicholson SE (2019) RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. Immunol Cell Biol 97:840–852
Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S (2002) Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3:196–200
Higgs R, Cormican P, Cahalane S, Allan B, Lloyd AT, Meade K, James T, Lynn DJ, Babiuk LA, O’Farrelly C (2006) Induction of a novel chicken Toll-like receptor following Salmonella enterica serovar Typhimurium infection. Infect Immun 74:1692–1698
Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, Crozat K, Sovath S, Han J, Beutler B (2003) Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 424:743–748
Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, Rixon FJ, Herold BC, Fitzgerald KA, Paludan SR (2012) Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol 13:737–743
Holm CK, Rahbek SH, Gad HH, Bak RO, Jakobsen MR, Jiang Z, Hansen AL, Jensen SK, Sun C, Thomsen MK, Laustsen A, Nielsen CG, Severinsen K, **ong Y, Burdette DL, Hornung V, Lebbink RJ, Duch M, Fitzgerald KA, Bahrami S, Mikkelsen JG, Hartmann R, Paludan SR (2016) Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat Commun 7:10680
Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777
Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G (2006) 5’-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997
Hron T, Pajer P, Pačes J, Bartůněk P, Elleder D (2015) Hidden genes in birds. Genome Biol 16:164
Hu X, Zou H, Qin A, Qian K, Shao H, Ye J (2016) Activation of Toll-like receptor 3 inhibits Marek’s disease virus infection in chicken embryo fibroblast cells. Arch Virol 161:521–528
Hua K, Li Y, Chen H, Ni J, Bi D, Luo R, ** H (2018) Functional characterization of duck TBK1 in IFN-β induction. Cytokine 111:325–333
Huang B, Qi ZT, Xu Z, Nie P (2010) Global characterization of interferon regulatory factor (IRF) genes in vertebrates: glimpse of the diversification in evolution. BMC Immunol 11:22
Huang Y, Temperley ND, Ren L, Smith J, Li N, Burt DW (2011) Molecular evolution of the vertebrate TLR1 gene family–a complex history of gene duplication, gene conversion, positive selection and co-evolution. BMC Evol Biol 11:149
Hur S (2019) Double-stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol 37:349–375
Ikeda T, Honjo K, Hirota Y, Onodera T (1993) Isolation of the chicken NF-kappa B p65 subunit-encoding cDNA and characterization of its products. Gene 133:237–242
Iqbal M, Philbin VJ, Smith AL (2005a) Expression patterns of chicken Toll-like receptor mRNA in tissues, immune cell subsets and cell lines. Vet Immunol Immunopathol 104:117–127
Iqbal M, Philbin VJ, Withanage GS, Wigley P, Beal RK, Goodchild MJ, Barrow P, McConnell I, Maskell DJ, Young J, Bumstead N, Boyd Y, Smith AL (2005b) Identification and functional characterization of chicken toll-like receptor 5 reveals a fundamental role in the biology of infection with Salmonella enterica serovar typhimurium. Infect Immun 73:2344–2350
Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678
Iwasaki A, Medzhitov R (2015) Control of adaptive immunity by the innate immune system. Nat Immunol 16:343–353
Janeway CA Jr (1989) Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54(Pt 1):1–13
Jax E, Müller I, Börno S, Borlinghaus H, Eriksson G, Fricke E, Timmermann B, Pendl H, Fiedler W, Klein K, Schreiber F, Wikelski M, Magor KE, Kraus RHS (2021) Health monitoring in birds using bio-loggers and whole blood transcriptomics. Sci Rep 11:10815
Jiao PR, Wei LM, Cheng YQ, Yuan RY, Han F, Liang J, Liu WL, Ren T, **n CA, Liao M (2012) Molecular cloning, characterization, and expression analysis of the Muscovy duck Toll-like receptor 3 (MdTLR3) gene. Poult Sci 91:2475–2481
** J, Ran JS, Yang CW, Jiang XS, Zhou YG, Feng ZQ, Wang Y, Lan D, Ren P, Liu YP (2017) Molecular characterization, expression, and functional analysis of chicken TRAF6. Genet Mol Res 16
Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, Lipford G, Bauer S (2002) Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol 3:499
Kaikai H, Zhao D, Liu Y, Liu Q, Huang X, Yang J, Zhang L, Li Y (2021) The E3 ubiquitin ligase TRIM25 inhibits Tembusu virus replication in vitro. Front Vet Sci 8:722113
Kang Y, Li Y, Yuan R, Feng M, **ang B, Sun M, Li Y, **e P, Tan Y, Ren T (2015) Host innate immune responses of ducks infected with Newcastle disease viruses of different pathogenicities. Front Microbiol 6:1283
Kanno A, Yamamoto C, Onji M, Fukui R, Saitoh S, Motoi Y, Shibata T, Matsumoto F, Muta T, Miyake K (2013) Essential role for Toll-like receptor 7 (TLR7)-unique cysteines in an intramolecular disulfide bond, proteolytic cleavage and RNA sensing. Int Immunol 25:413–422
Karin M (1999) How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18:6867–6874
Karpala AJ, Lowenthal JW, Bean AG (2008) Activation of the TLR3 pathway regulates IFNbeta production in chickens. Dev Comp Immunol 32:435–444
Karpala AJ, Stewart C, McKay J, Lowenthal JW, Bean AG (2011) Characterization of chicken Mda5 activity: regulation of IFN-beta in the absence of RIG-I functionality. J Immunol 186:5397–5405
Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S (2008) Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610
Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105
Kato K, Ahmad S, Zhu Z, Young JM, Mu X, Park S, Malik HS, Hur S (2021) Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol Cell 81:599-613.e8
Kawai T, Akira S (2006a) Innate immune recognition of viral infection. Nat Immunol 7:131–137
Kawai T, Akira S (2006b) TLR signaling. Cell Death Differ 13:816–825
Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384
Kawasaki T, Kawai T (2014) Toll-like Receptor Signaling Pathways Front Immunol 5:461
Keestra AM, de Zoete MR, Bouwman LI, Vaezirad MM, van Putten JP (2013) Unique features of chicken Toll-like receptors. Dev Comp Immunol 41:316–323
Keestra AM, de Zoete MR, Bouwman LI, van Putten JP (2010) Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J Immunol 185:460–467
Keestra AM, van Putten JP (2008) Unique properties of the chicken TLR4/MD-2 complex: selective lipopolysaccharide activation of the MyD88-dependent pathway. J Immunol 181:4354–4362
Kim TH, Kern C, Zhou H (2020) Knockout of IRF7 highlights its modulator function of host response against avian influenza virus and the involvement of MAPK and TOR signaling pathways in chicken. Genes (Basel) 11
Kim TH, Zhou H (2015) Functional analysis of chicken IRF7 in response to dsRNA analog poly(I:C) by integrating overexpression and knockdown. PLoS One 10:e0133450
Kogut MH, Iqbal M, He H, Philbin V, Kaiser P, Smith A (2005) Expression and function of Toll-like receptors in chicken heterophils. Dev Comp Immunol 29:791–807
Kogut MH, Swaggerty C, He H, Pevzner I, Kaiser P (2006) Toll-like receptor agonists stimulate differential functional activation and cytokine and chemokine gene expression in heterophils isolated from chickens with differential innate responses. Microbes Infect 8:1866–1874
Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M (2003) Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg Infect Dis 9:311–322
Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S (2011) Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147:423–435
Lang X, Tang T, ** T, Ding C, Zhou R, Jiang W (2017) TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med 214:459–473
Le Goffic R, Balloy V, Lagranderie M, Alexopoulou L, Escriou N, Flavell R, Chignard M, Si-Tahar M (2006) Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog 2:e53
Lee SB, Park YH, Chungu K, Woo SJ, Han ST, Choi HJ, Rengaraj D, Han JY (2020) Targeted knockout of MDA5 and TLR3 in the DF-1 chicken fibroblast cell line impairs innate immune response against RNA ligands. Front Immunol 11:678
Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM (2008) The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A 105:258–263
Li H, Zhai Y, Fan Y, Chen H, Zhang A, ** H, Luo R (2016) Molecular cloning and functional characterization of duck mitochondrial antiviral-signaling protein (MAVS). Dev Comp Immunol 56:1–6
Li K, Liu Y, Xu Z, Zhang Y, Luo D, Gao Y, Qian Y, Bao C, Liu C, Zhang Y, Qi X, Cui H, Wang Y, Gao L, Wang X (2019) Avian oncogenic herpesvirus antagonizes the cGAS-STING DNA-sensing pathway to mediate immune evasion. PLoS Pathog 15:e1007999
Li Y, Ma L, Rao Z, Zhou P, Zheng H, Luo R (2021) Characterization of duck IκB kinase β involved in innate immunity. Dev Comp Immunol 125:104208
Lin R, Heylbroeck C, Pitha PM, Hiscott J (1998) Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18:2986–2996
Liniger M, Summerfield A, Zimmer G, McCullough KC, Ruggli N (2012) Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. J Virol 86:705–717
Liu G, Park HS, Pyo HM, Liu Q, Zhou Y (2015a) Influenza A virus panhandle structure is directly involved in RIG-I activation and interferon induction. J Virol 89:6067–6079
Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR (2008) Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 320:379–381
Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ (2015b) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630
Liu YJ (2005) IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Ann Rev Immunol 23:275–306
Loo YM, Gale M Jr (2011) Immune signaling by RIG-I-like receptors. Immunity 34:680–692
Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA 101:5598–5603
Ma Z, Damania B (2016) The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19:150–158
MacDonald MR, **a J, Smith AL, Magor KE (2008) The duck toll like receptor 7: genomic organization, expression and function. Mol Immunol 45:2055–2061
Magor KE, Miranzo-Navarro D, Barber MRW, Petkau K, Fleming-Canepa X, Blyth GAD, Blaine AH (2013) Defense genes missing from the flight division. Dev Comp Immunol 41:377–388
Maillard PV, van der Veen AG, Poirier EZ, Reis e Sousa C (2019) Slicing and dicing viruses: antiviral RNA interference in mammals. EMBO J 38:e100941
McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T (2004) IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A 101:233–238
Meyerson NR, Zhou L, Guo YR, Zhao C, Tao YJ, Krug RM, Sawyer SL (2017) Nuclear TRIM25 specifically targets influenza virus ribonucleoproteins to block the onset of RNA chain elongation. Cell Host Microbe 22:627-638.e7
Miranzo-Navarro D, Magor KE (2014) Activation of duck RIG-I by TRIM25 is independent of anchored ubiquitin. PLoS ONE 9:e86968
Morris KM, Hindle MM, Boitard S, Burt DW, Danner AF, Eory L, Forrest HL, Gourichon D, Gros J, Hillier LW, Jaffredo T, Khoury H, Lansford R, Leterrier C, Loudon A, Mason AS, Meddle SL, Minvielle F, Minx P, Pitel F, Seiler JP, Shimmura T, Tomlinson C, Vignal A, Webster RG, Yoshimura T, Warren WC, Smith J (2020) The quail genome: insights into social behaviour, seasonal biology and infectious disease response. BMC Biol 18:14
Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ (2009) An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem 284:25404–25411
Nagy N, Bódi I, Oláh I (2016) Avian dendritic cells: phenotype and ontogeny in lymphoid organs. Dev Comp Immunol 58:47–59
Nawab A, An L, Wu J, Li G, Liu W, Zhao Y, Wu Q, **ao M (2019) Chicken toll-like receptors and their significance in immune response and disease resistance. Int Rev Immunol 38:284–306
Neerukonda SN, Katneni U (2020) Avian pattern recognition receptor sensing and signaling. Vet Sci 7
Nistal-Villan E, Gack MU, Martinez-Delgado G, Maharaj NP, Inn KS, Yang H, Wang R, Aggarwal AK, Jung JU, Garcia-Sastre A (2010) Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem 285:20252–20261
Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus AD, Fouchier RA (2006) Global patterns of influenza A virus in wild birds. Science 312(5772):384-388
Oven I, Resman Rus K, Dusanic D, Bencina D, Keeler CL Jr, Narat M (2013) Diacylated lipopeptide from Mycoplasma synoviae mediates TLR15 induced innate immune responses. Vet Res 44:99
Pantin-Jackwood M, Swayne DE, Smith D, Shepherd E (2013) Effect of species, breed and route of virus inoculation on the pathogenicity of H5N1 highly pathogenic influenza (HPAI) viruses in domestic ducks. Vet Res 44:62
Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, Hohng S, Hur S (2012) Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci U S A 109:E3340–E3349
Philbin VJ, Iqbal M, Boyd Y, Goodchild MJ, Beal RK, Bumstead N, Young J, Smith AL (2005) Identification and characterization of a functional, alternatively spliced Toll-like receptor 7 (TLR7) and genomic disruption of TLR8 in chickens. Immunology 114:507–521
Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU (2012) Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8:e1003059
Ranaware PB, Mishra A, Vijayakumar P, Gandhale PN, Kumar H, Kulkarni DD, Raut AA (2016) Genome wide host gene expression analysis in chicken lungs infected with avian influenza viruses. PLoS One 11:e0153671
Raven N, Lisovski S, Klaassen M, Lo N, Madsen T, Ho SYW, Ujvari B (2017) Purifying selection and concerted evolution of RNA-sensing toll-like receptors in migratory waders. Infect Genet Evol 53:135–145
Rehman MS, Rehman SU, Yousaf W, Hassan FU, Ahmad W, Liu Q, Pan H (2021) The potential of toll-like receptors to modulate avian immune system: exploring the effects of genetic variants and phytonutrients. Front Genet 12:671235
Rehwinkel J, Gack MU (2020) RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20:537–551
Ren Z, Ding T, Zuo Z, Xu Z, Deng J, Wei Z (2020) Regulation of MAVS expression and signaling function in the antiviral innate immune response. Front Immunol 11:1030
Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A (2005) The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci U S A 102:9577–9582
Rosadini CV, Kagan JC (2015) Microbial strategies for antagonizing Toll-like-receptor signal transduction. Curr Opin Immunol 32:61–70
Saiz JC, Martín-Acebes MA, Blázquez AB, Escribano-Romero E, Poderoso T, Jiménez de Oya N (2021) Pathogenicity and virulence of West Nile virus revisited eight decades after its first isolation. Virulence 12:1145–1173
Santhakumar D, Rubbenstroth D, Martinez-Sobrido L, Munir M (2017) Avian Interferons and Their Antiviral Effectors. Front Immunol 8:49
Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, Hartmann G (2009a) Approaching the RNA ligand for RIG-I? Immunol Rev 227:66–74
Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G (2009b) Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34
Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, García-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM (2014) Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695
Schwarz H, Schneider K, Ohnemus A, Lavric M, Kothlow S, Bauer S, Kaspers B, Staeheli P (2007) Chicken toll-like receptor 3 recognizes its cognate ligand when ectopically expressed in human cells. J Interferon Cytokine Res 27:97–101
Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682
Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J (2003) Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151
Sick C, Schultz U, Staeheli P (1996) A family of genes coding for two serologically distinct chicken interferons. J Biol Chem 271:7635–7639
St Paul M, Brisbin JT, Abdul-Careem MF, Sharif S (2013) Immunostimulatory properties of Toll-like receptor ligands in chickens. Vet Immunol Immunopathol 152:191–199
Sun L, Wu J, Du F, Chen X, Chen ZJ (2013a) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791
Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ (2006) The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24:633–642
Sun Y, Ding N, Ding SS, Yu S, Meng C, Chen H, Qiu X, Zhang S, Yu Y, Zhan Y, Ding C (2013b) Goose RIG-I functions in innate immunity against Newcastle disease virus infections. Mol Immunol 53:321–327
Sun Y, Mao X, Zheng H, Wu W, Rehman ZU, Liao Y, Meng C, Qiu X, Tan L, Song C, Xu L, Yu S, Ding C (2019) Goose MAVS functions in RIG-I-mediated IFN-β signaling activation. Dev Comp Immunol 93:58–65
Świderská Z, Šmídová A, Buchtová L, Bryjová A, Fabiánová A, Munclinger P, Vinkler M (2018) Avian Toll-like receptor allelic diversity far exceeds human polymorphism: an insight from domestic chicken breeds. Sci Rep 8:17878
Te Velthuis AJW, Long JC, Bauer DLV, Fan RLY, Yen HL, Sharps J, Siegers JY, Killip MJ, French H, Oliva-Martin MJ, Randall RE, de Wit E, van Riel D, Poon LLM, Fodor E (2018) Mini viral RNAs act as innate immune agonists during influenza virus infection. Nat Microbiol 3:1234–1242
Temperley ND, Berlin S, Paton IR, Griffin DK, Burt DW (2008) Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics 9:62
van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche-Grass S, Borg A, Kjær S, Snijders AP, Reis e Sousa C (2018) The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. Embo j 37
Velova H, Gutowska-Ding MW, Burt DW, Vinkler M (2018) Toll-like receptor evolution in birds: gene duplication, pseudogenization, and diversifying selection. Mol Biol Evol 35:2170–2184
Vinkler M, Bainová H, Bryja J (2014) Protein evolution of Toll-like receptors 4, 5 and 7 within Galloanserae birds. Genet Sel Evol 46:72
Volmer C, Soubies SM, Grenier B, Guerin JL, Volmer R (2011) Immune response in the duck intestine following infection with low-pathogenic avian influenza viruses or stimulation with a Toll-like receptor 7 agonist administered orally. J Gen Virol 92:534–543
Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412:346–351
Wang S, Yu M, Liu A, Bao Y, Qi X, Gao L, Chen Y, Liu P, Wang Y, **ng L, Meng L, Zhang Y, Fan L, Li X, Pan Q, Zhang Y, Cui H, Li K, Liu C, He X, Gao Y, Wang X (2021) TRIM25 inhibits infectious bursal disease virus replication by targeting VP3 for ubiquitination and degradation. PLoS Pathog 17:e1009900
Wang Y, Yin Y, Lan X, Ye F, Tian K, Zhao X, Yin H, Li D, Xu H, Liu Y, Zhu Q (2017) Molecular characterization, expression of chicken TBK1 gene and its effect on IRF3 signaling pathway. PLoS One 12:e0177608
Weber M, Sediri H, Felgenhauer U, Binzen I, Banfer S, Jacob R, Brunotte L, Garcia-Sastre A, Schmid-Burgk JL, Schmidt T, Hornung V, Kochs G, Schwemmle M, Klenk HD, Weber F (2015) Influenza virus adaptation PB2-627K modulates nucleocapsid inhibition by the pathogen sensor RIG-I. Cell Host Microbe 17:309–319
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179
Wei L, Cui J, Song Y, Zhang S, Han F, Yuan R, Gong L, Jiao P, Liao M (2014) Duck MDA5 functions in innate immunity against H5N1 highly pathogenic avian influenza virus infections. Vet Res 45:66
Wei L, Jiao P, Song Y, Cao L, Yuan R, Gong L, Cui J, Zhang S, Qi W, Yang S, Liao M (2013) Host immune responses of ducks infected with H5N1 highly pathogenic avian influenza viruses of different pathogenicities. Vet Microbiol 166:386–393
Wei X, Qian W, Sizhu S, Shi L, ** M, Zhou H (2016) Molecular cloning and functional analysis of the duck TIR domain-containing adaptor inducing IFN-β (TRIF) gene. Dev Comp Immunol 65:369–376
Wheaton S, Lambourne MD, Sarson AJ, Brisbin JT, Mayameei A, Sharif S (2007) Molecular cloning and expression analysis of chicken MyD88 and TRIF genes. DNA Seq 18:480–486
Wille M, Geoghegan JL, Holmes EC (2021) How accurately can we assess zoonotic risk? PLoS Biol 19:e3001135
Wille M, Shi M, Klaassen M, Hurt AC, Holmes EC (2019) Virome heterogeneity and connectivity in waterfowl and shorebird communities. Isme j 13:2603–2616
Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S (2013) Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152:276–289
Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S (2014) Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell 55:511–523
Wu J, Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488
Wu Z, Rothwell L, Young JR, Kaufman J, Butter C, Kaiser P (2010) Generation and characterization of chicken bone marrow-derived dendritic cells. Immunology 129:133–145
Wu Z, Zhang W, Wu Y, Wang T, Wu S, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Pan L, Merits A, Chen S, Cheng A (2019) Binding of the duck tembusu virus protease to STING is mediated by NS2B and is crucial for STING cleavage and for impaired induction of IFN-β. J Immunol 203:3374–3385
**ao Y, Evseev D, Stevens CA, Moghrabi A, Miranzo-Navarro D, Fleming-Canepa X, Tetrault DG, Magor KE (2020) Influenza PB1-F2 inhibits avian MAVS signaling. Viruses 12:e409
**ao Y, Reeves MB, Caulfield AF, Evseev D, Magor KE (2018) The core promoter controls basal and inducible expression of duck retinoic acid inducible gene-I (RIG-I). Mol Immunol 103:156–165
Xu L, Yu D, Fan Y, Liu YP, Yao YG (2019) Evolutionary selection on MDA5 and LGP2 in the chicken preserves antiviral competence in the absence of RIG-I. J Genet Genomics 46:499–503
Xu W, Shao Q, Zang Y, Guo Q, Zhang Y, Li Z (2015) Pigeon RIG-I function in innate immunity against H9N2 IAV and IBDV. Viruses 7:4131–4151
Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S (2003) Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640–643
Yang J, Zhou M, Zhong Y, Xu L, Zeng C, Zhao X, Zhang M (2021) Gene duplication and adaptive evolution of Toll-like receptor genes in birds. Dev Comp Immunol 119:103990
Yilmaz A, Shen S, Adelson DL, Xavier S, Zhu JJ (2005) Identification and sequence analysis of chicken Toll-like receptors. Immunogenetics 56:743–753
Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ (2010) Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–330
Zhai Y, Luo F, Chen Y, Zhou S, Li Z, Liu M, Bi D, ** H (2015) Molecular characterization and functional analysis of duck TRAF6. Dev Comp Immunol 49:1–6
Zhang M, Song K, Li C, Chen Z, Ding C, Liu G (2015) Molecular cloning of Peking duck Toll-like receptor 3 (duTLR3) gene and its responses to reovirus infection. Virol J 12:207
Zhang P, Ding Z, Liu X, Chen Y, Li J, Tao Z, Fei Y, Xue C, Qian J, Wang X, Li Q, Stoeger T, Chen J, Bi Y, Yin R (2018a) Enhanced replication of virulent Newcastle disease virus in chicken macrophages is due to polarized activation of cells by inhibition of TLR7. Front Immunol 9:366
Zhang W, Jiang B, Zeng M, Duan Y, Wu Z, Wu Y, Wang T, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Pan L, Chen S, Cheng A (2020) Binding of duck Tembusu virus nonstructural protein 2A to duck STING disrupts induction of its signal transduction cascade to inhibit beta interferon induction. J Virol 94
Zhang Z, Ohto U, Shibata T, Krayukhina E, Taoka M, Yamauchi Y, Tanji H, Isobe T, Uchiyama S, Miyake K, Shimizu T (2016) Structural analysis reveals that Toll-like receptor 7 is a dual receptor for guanosine and single-stranded RNA. Immunity 45:737–748
Zhang Z, Ohto U, Shibata T, Taoka M, Yamauchi Y, Sato R, Shukla NM, David SA, Isobe T, Miyake K, Shimizu T (2018b) Structural analyses of toll-like receptor 7 reveal detailed RNA sequence specificity and recognition mechanism of agonistic ligands. Cell Rep 25:3371-3381.e5
Zheng W, Satta Y (2018) Functional Evolution of Avian RIG-I-Like Receptors. Genes (Basel) 9
Zhou JR, Liu JH, Li HM, Zhao Y, Cheng Z, Hou YM, Guo HJ (2020) Regulatory effects of chicken TRIM25 on the replication of ALV-A and the MDA5-mediated type I interferon response. Vet Res 51:145
Zhou P, Zeng Y, Rao Z, Li Y, Zheng H, Luo R (2021) Molecular characterization and functional analysis of duck IKKα. Dev Comp Immunol 115:103880
Zhu J, Xu S, Li X, Wang J, Jiang Y, Hu W, Ruan W (2020) Infectious bronchitis virus inhibits activation of the TLR7 pathway, but not the TLR3 pathway. Arch Virol 165:2037–2043
Acknowledgements
I thank Lee K. Campbell and two anonymous reviewers for critical reading of the manuscript and past and present members of the lab for research contributing to our understanding of avian pattern recognition receptors and viral antagonism.
Funding
Research in my laboratory is funded by an NSERC Discovery grant from the Natural Sciences and Engineering Council of Canada and the Canadian Institutes of Health Research PJT 159442.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Magor, K.E. Evolution of RNA sensing receptors in birds. Immunogenetics 74, 149–165 (2022). https://doi.org/10.1007/s00251-021-01238-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01238-1