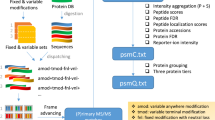
Abstract
In recent years the organization of mass spectrometry (MS) data obtained in large-scale proteomics projects became an important issue. This has catalyzed the development of a few different database schemes for storing MS data, as well as some dedicated user interfaces. However, many of these projects are still rather immature and often do not cover all needs. Because our needs were quite specific, it was necessary to build a database that accommodates all the major types of experiments generated in house and that could be easily extended by new modules made by collaborators or students. A database application named “YassDB” will be described in this chapter. The application is implemented in a “three-tier” application architecture, with a database layer, a middle layer consisting of web services and a client layer, containing the user interface. This offers high flexibility: it allows other applications, written in any language, to be written as clients to the database. The setup and use of the YassDB database application with two client programs “pProRep” and “VEMS” will be outlined.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Orchard, S., Hermjakob, H., Julian, R. K., Jr., et al. (2004) Common interchange standards for proteomics data: public availability of tools and schema. Proteomics 4, 490–491.
Garwood, K., McLaughlin, T., Garwood, C., et al. (2004) PEDRo: a database for storing, searching and disseminating experimental proteomics data. BMC Genomics 5, 68.
Patterson, S. D. and Aebersold, R. H. (2003) Proteomics: the first decade and beyond. Nat. Genet. 33, 311–323.
Taylor, C. F., Paton, N. W., Garwood, K. L., et al. (2003) A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nature Biotech. 21, 247–254.
Garden, P., Alm, R., and Hakkinen, J. (2005) PROTEIOS: an open source proteomics initiative. Bioinformatics 21, 2085–2087.
Matthiesen, R., Lundsgaard, M., Welinder, K. G., and Bauw, G. (2003) Interpreting peptide mass spectra by VEMS. Bioinformatics 19, 792–793.
Matthiesen, R., Bunkenborg, J., Stensballe, A., Jensen, O. N., Welinder, K. G., and Bauw, G. (2004) Database-independent, database-dependent, and extended interpretation of peptide mass spectra in VEMS V2.0. Proteomics 4, 2583–2593.
Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 18, 3551–3567.
Ramakrishnan, R. and Gehrke, J. (2003) Database Management Systems. McGraw-Hill, New York.
Deitel, H. M., Deitel, P. J., Gadzik, J. P., Lomelí, K., Santry, S. E., and Zhang, S. (2003) Java Web Services for Experienced Programmers. Prentice Hall, Upper Saddle River, NJ.
Chopra, V., Bakore, A., Eaves, J., Galbraith, B., Li, S., and Wiggers, C. (2004) Professional Apache Tomcat 5. Wrox, Hoboken, NJ.
Converse, T. and Park, J. (2004) PHP5 and MySQL Bible. Wiley, Hoboken, NJ.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2007 Humana Press Inc., Totowa, NJ
About this protocol
Cite this protocol
Thomsen, A.L., Laukens, K., Matthiesen, R., Jensen, O.N. (2007). Organization of Proteomics Data With YassDB. In: Matthiesen, R. (eds) Mass Spectrometry Data Analysis in Proteomics. Methods in Molecular Biology, vol 367. Humana Press. https://doi.org/10.1385/1-59745-275-0:271
Download citation
DOI: https://doi.org/10.1385/1-59745-275-0:271
Publisher Name: Humana Press
Print ISBN: 978-1-58829-563-7
Online ISBN: 978-1-59745-275-5
eBook Packages: Springer Protocols