Abstract
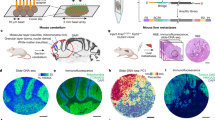
The Visium Spatial Gene Expression Solution (Visium 10×) allows for the mRNA analysis using high throughput sequencing and maps a transcriptional expression pattern in tissue sections using high-resolution microscope imaging in ex-vivo human and mice samples. The workflow surveys spatial global gene expression in tissue sections, exploiting the whole transcriptome profiling and defining the set of transcripts via targeted gene panels. An automated cell type annotation allows a comparison with control tissue samples. This technique delineates cancerous or diseased tissue boundaries and details gene expression gradients in the tissue surrounding the tumor or pathologic nests. Remarkably, the Visium 10× allows for whole transcriptome and targeted analysis without the loss of spatial information. This approach provides gene expression data within the context of tissue architecture, tissue microenvironments, and cell groups. It can be used in association with therapy, anti-angiogenic therapy, and immunotherapy to improve treatment response.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Baghban R, Roshangar L, Jahanban-Esfahlan R et al (2020) Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal 18:59. https://doi.org/10.1186/s12964-020-0530-4
Marquardt A, Solimando AG, Kerscher A et al (2021) Subgroup-independent map** of renal cell carcinoma-machine learning reveals prognostic mitochondrial gene signature beyond histopathologic boundaries. Front Oncol 11:621278. https://doi.org/10.3389/fonc.2021.621278
Solimando AG, Da Vià MC, Leone P et al (2021) Halting the vicious cycle within the multiple myeloma ecosystem: blocking JAM-A on bone marrow endothelial cells restores Angiogenic homeostasis and suppresses tumor progression. Haematologica 106:1943–1956. https://doi.org/10.3324/haematol.2019.239913
Solimando AG, Summa SD, Vacca A et al (2020) Cancer-associated angiogenesis: the endothelial cell as a checkpoint for immunological patrolling. Cancers (Basel) 12:E3380. https://doi.org/10.3390/cancers12113380
Da Vià MC, Solimando AG, Garitano-Trojaola A et al (2020) CIC mutation as a molecular mechanism of acquired resistance to combined BRAF-MEK inhibition in extramedullary multiple myeloma with central nervous system involvement. Oncologist 25:112–118. https://doi.org/10.1634/theoncologist.2019-0356
Solimando AG, Da Vià MC, Cicco S et al (2019) High-risk multiple myeloma: integrated clinical and omics approach dissects the neoplastic clone and the tumor microenvironment. J Clin Med 8:997. https://doi.org/10.3390/jcm8070997
Georganaki M, van Hooren L, Dimberg A (2018) Vascular targeting to increase the efficiency of immune checkpoint blockade in cancer. Front Immunol 9:3081. https://doi.org/10.3389/fimmu.2018.03081
Dimopoulos MA, Moreau P, Terpos E et al (2022) Corrigendum to “multiple myeloma: EHA-ESMO clinical practice guidelines for diagnosis, treatment and follow-up”: [Ann Oncol 2021; 32(3): 309–322]. Ann Oncol 33:117. https://doi.org/10.1016/j.annonc.2021.10.001
Bolli N, Avet-Loiseau H, Wedge DC et al (2014) Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 5:2997. https://doi.org/10.1038/ncomms3997
Palumbo A, Avet-Loiseau H, Oliva S et al (2015) Revised international staging system for multiple myeloma: a report from international myeloma working group. J Clin Oncol 33:2863–2869. https://doi.org/10.1200/JCO.2015.61.2267
Berglund E, Maaskola J, Schultz N et al (2018) Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun 9:2419. https://doi.org/10.1038/s41467-018-04724-5
Krebs M, Solimando AG, Kalogirou C et al (2020) MiR-221-3p regulates VEGFR2 expression in high-risk prostate cancer and represents an escape mechanism from sunitinib in vitro. J Clin Med 9:E670. https://doi.org/10.3390/jcm9030670
Lv J, Shi Q, Han Y et al (2021) Spatial transcriptomics reveals gene expression characteristics in invasive micropapillary carcinoma of the breast. Cell Death Dis 12:1095. https://doi.org/10.1038/s41419-021-04380-6
Shadbad MA, Safaei S, Brunetti O et al (2021) A systematic review on the therapeutic potentiality of PD-L1-inhibiting MicroRNAs for triple-negative breast cancer: toward single-cell sequencing-guided biomimetic delivery. Genes (Basel) 12:1206. https://doi.org/10.3390/genes12081206
Stark R, Grzelak M, Hadfield J (2019) RNA sequencing: the teenage years. Nat Rev Genet 20:631–656. https://doi.org/10.1038/s41576-019-0150-2
Ståhl PL, Salmén F, Vickovic S et al (2016) Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353:78–82. https://doi.org/10.1126/science.aaf2403
Liyanage S, Dassanayake RS, Bouyanfif A et al (2017) Optimization and validation of cryostat temperature conditions for trans-reflectance mode FTIR microspectroscopic imaging of biological tissues. MethodsX 4:118–127. https://doi.org/10.1016/j.mex.2017.01.006
Joglekar A, Prjibelski A, Mahfouz A et al (2021) A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat Commun 12:463. https://doi.org/10.1038/s41467-020-20343-5
Rao L, Giannico D, Leone P et al (2020) HB-EGF-EGFR signaling in bone marrow endothelial cells mediates angiogenesis associated with multiple myeloma. Cancers (Basel) 12:E173. https://doi.org/10.3390/cancers12010173
Misra A, Baker CD, Pritchett EM et al (2021) Characterizing neonatal heart maturation, regeneration, and scar resolution using spatial transcriptomics. J Cardiovasc Dev Dis 9:1. https://doi.org/10.3390/jcdd9010001
Brandl A, Solimando A, Mokhtari Z et al (2021) Junctional adhesion molecule-C expression specifies a CD138low/Neg multiple myeloma cell population in mice and humans. Blood Adv. https://doi.org/10.1182/bloodadvances.2021004354
Hafemeister C, Satija R (2019) Normalization and variance stabilization of single-cell RNA-Seq data using regularized negative binomial regression. Genome Biol 20:296. https://doi.org/10.1186/s13059-019-1874-1
Nicole M, Anderson M. Celeste, Simon (2020) The tumor microenvironment. Current Biology 30(16):R921–R925 S0960982220309337 https://doi.org/10.1016/j.cub.2020.06.081
Mark, Spaw Shrikant, Anant Sufi Mary, Thomas (2017) Stromal contributions to the carcinogenic process. Molecular Carcinogenesis 56(4):1199–1213. https://doi.org/10.1002/mc.22583
Gnoni, Antonio Brunetti, Oronzo Longo, Vito Calabrese, Angela Argentiero, Antonel-la Calbi, Roberto Solimando Antonio, Giovanni Licchetta, Antonella (2020) Immune system and bone microenvironment: rationale for targeted cancer therapies. Oncotarget 11(4):480–487. https://doi.org/10.18632/oncotarget.27439
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Solimando, A.G., Desantis, V., Da Vià, M.C. (2023). Visualizing the Interactions Sha** the Imaging of the Microenvironment in Human Cancers. In: Ribatti, D. (eds) Tumor Angiogenesis Assays. Methods in Molecular Biology, vol 2572. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2703-7_5
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2703-7_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2702-0
Online ISBN: 978-1-0716-2703-7
eBook Packages: Springer Protocols