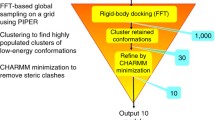
Abstract
Many of the biological functions of the cell are driven by protein–protein interactions. However, determining which proteins interact and exactly how they do so to enable their functions, remain major research questions. Functional interactions are dependent on a number of complicated factors; therefore, modeling the three-dimensional structure of protein–protein complexes is still considered a complex endeavor. Nevertheless, the rewards for modeling protein interactions to atomic level detail are substantial, and there are numerous examples of how models can provide useful information for drug design, protein engineering, systems biology, and understanding of the immune system. Here, we provide practical guidelines for docking proteins using the web-server, SwarmDock, a flexible protein–protein docking method. Moreover, we provide an overview of the factors that need to be considered when deciding whether docking is likely to be successful.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Tovchigrechko A, Vakser IA (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res 34:W310–W314
Lyskov S, Gray JJ (2008) The RosettaDock server for local protein-protein docking. Nucleic Acids Res 36:W233–W238
Garzon JI et al (2009) FRODOCK: a new approach for fast rotational protein-protein docking. Bioinformatics 25(19):2544–2551
Macindoe G, Mavridis L, Venkatraman V, Devignes M-D, Ritchie DW (2010) HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 38:W445–W449
Mashiach E et al (2010) An integrated suite of fast docking algorithms. Proteins 78(15):3197–3204
Huang S-Y, Zou X (2010) MDockPP: a hierarchical approach for protein-protein docking and its application to CAPRI rounds 15-19. Proteins 78(15):3096–3103
Pierce BG, Hourai Y, Weng Z (2011) Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One 6(9):e24657
Jiménez-García B, Pons C, Fernández-Recio J (2013) pyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics 29(13):1698–1699
van Zundert GCP, Bonvin AMJJ (2014) Modeling protein-protein complexes using the HADDOCK webserver “modeling protein complexes with HADDOCK”. Methods Mol Biol 1137:163–179
Viswanath S, Ravikant DVS, Elber R (2014) DOCK/PIERR: web server for structure prediction of protein-protein complexes. Methods Mol Biol 1137:199–207
Esquivel-Rodriguez J, Filos-Gonzalez V, Li B, Kihara D (2014) Pairwise and multimeric protein-protein docking using the LZerD program suite. Methods Mol Biol 1137:209–234
de Vries SJ, Schindler CEM, Chauvot de Beauchêne I, Zacharias M (2015) A web interface for easy flexible protein-protein docking with ATTRACT. Biophys J 108(3):462–465
Kozakov D et al (2017) The ClusPro web server for protein-protein docking. Nat Protoc 12(2):255–278
Lee H, Seok C (2017) Template-based prediction of protein-peptide interactions by using GalaxyPepDock. Methods Mol Biol 1561:37–47
Moal IH, Bates PA (2010) SwarmDock and the use of normal modes in protein-protein docking. Int J Mol Sci 11(10):3623–3648
Li X, Moal IH, Bates PA (2010) Detection and refinement of encounter complexes for protein–protein docking: taking account of macromolecular crowding. Proteins 8(15):3189–3196
Torchala M, Moal IH, Chaleil RA, Fernandez-Recio J, Bates PA (2013) SwarmDock: a server for flexible protein-protein docking. Bioinformatics 29(6):807–809
Torchala M, Bates PA (2014) Predicting the structure of protein-protein complexes using the SwarmDock web server. Methods Mol Biol 1137:181–197
Vajda S, Hall DR, Kozakov D (2013) Sampling and scoring: a marriage made in heaven. Proteins 81(11):1874–1884
Moal IH, Torchala M, Bates PA, Fernandez-Recio J (2013) The scoring of poses in protein-protein docking: current capabilities and future directions. BMC Bioinformatics 14:286
Barradas-Bautista D, Moal IH, Fernández-Recio J (2017) A systematic analysis of scoring functions in rigid-body protein docking: the delicate balance between the predictive rate improvement and the risk of overtraining. Proteins 85(7):1287–1297
Hayes TW, Moal IH (2017) Modeling protein conformational transition pathways using collective motions and the LASSO method. J Chem Theory Comput 13(3):1401–1410
Liu S, Zhang C, Zhou H, Zhou Y (2004) A physical reference state unifies the structure-derived potential of mean force for protein folding and binding. Proteins 56(1):93–101
Kennedy J, Eberhart R (1995) Particle swarm optimization. IEEE International Conference on Neural Networks, Perth
Solis FJ, Wets RJ-B (1981) Minimization by random search techniques. Math Oper Res 6(1):19–30
Tobi D (2010) Designing coarse grained-and atom based-potentials for protein-protein docking. BMC Struct Biol 10:40
Torchala M, Moal IH, Chaleil RA, Agius R, Bates PA (2013) A Markov-chain model description of binding funnels to enhance the ranking of docked solutions. Proteins 81(12):2143–2149
Moal IH et al (2017) IRaPPA: information retrieval based integration of biophysical models for protein assembly selection. Bioinformatics 33(12):1806–1813
Dobbins SE, Lesk VI, Sternberg MJE (2008) Insights into protein flexibility: the relationship between normal modes and conformational change upon protein-protein docking. Proc Natl Acad Sci U S A 105(30):10390–10395
Karaca E, Bonvin AMJJ (2011) A multidomain flexible docking approach to deal with large conformational changes in the modeling of biomolecular complexes. Structure 19(4):555–565
Marsh JA, Teichmann SA (2011) Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure 19(6):859–867
Chen H, Sun Y, Shen Y (2017) Predicting protein conformational changes for unbound and homology docking: learning from intrinsic and induced flexibility. Proteins 85(3):544–556
Wang Q, Canutescu AA, Dunbrack RL (2008) SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nat Protoc 3(12):1832–1847
Soto CS, Fasnacht M, Zhu J, Forrest L, Honig B (2008) Loop modeling: sampling, filtering, and scoring. Proteins 70(3):834–843
Brooks BR et al (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30(10):1545–1614
Suhre K, Sanejouand Y-H (2004) ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res 32:W610–W614
Engelbrecht AP (2005) Fundamentals of computational swarm intelligence. Wiley, Hoboken, NJ
Moal IH, Jimenez-Garcia B, Fernandez-Recio J (2015) CCharPPI web server: computational characterization of protein-protein interactions from structure. Bioinformatics 31(1):123–125
Pfeiffenberger E, Chaleil RAG, Moal IH, Bates PA (2017) A machine learning approach for ranking clusters of docked protein-protein complexes by pairwise cluster comparison. Proteins 85(3):528–543
van Zundert GCP et al (2016) The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol 428(4):720–725
Svergun DI et al (1998) Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc Natl Acad Sci U S A 95(5):2267–2272
Svergun D, Barberato C, Koch MHJ (1995) CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28(6):768–773
Shvartsburg AA, Jarrold MF (1996) An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem Phys Lett 261(1–2):86–91
Lasker K, Sali A, Wolfson HJ (2010) Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins 78(15):3205–3211
Russel D et al (2012) Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol 10(1):e1001244
Moal IH, Fernández-Recio J (2012) SKEMPI: a structural kinetic and energetic database of mutant protein interactions and its use in empirical models. Bioinformatics 28(20):2600–2607
Fowler DM, Fields S (2014) Deep mutational scanning: a new style of protein science. Nat Methods 11(8):801–807
Andreani J, Faure G, Guerois R (2012) Versatility and invariance in the evolution of homologous heteromeric interfaces. PLoS Comput Biol 8(8):e1002677
Nadaradjane AA, Guerois R, Andreani J (2018) Protein-protein docking using evolutionary information. Methods Mol Biol 1764:429–447
Reichmann D et al (2005) The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci U S A 102(1):57–62
Jankauskaite J, Jimenez-Garcia B, Dapkunas J, Fernandez-Recio J, Moal IH (2019) SKEMPI 2.0: an updated benchmark of changes in protein-protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics 35(3):462–469
Melandri D et al (2018) The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat Immunol 19(12):1352–1365
Palakodeti A et al (2012) The molecular basis for modulation of human Vγ9Vδ2 T cell responses by CD277/butyrophilin-3 (BTN3A)-specific antibodies. J Biol Chem 287(39):32780–32790
Willcox BE, Willcox CR (2019) γδ TCR ligands: the quest to solve a 500-million-year-old mystery. Nat Immunol 20(2):121–128
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Acknowledgments
This work was supported by the European Molecular Biology Laboratory [IHM], the Biotechnology and Biological Sciences Research Council [Future Leader Fellowship BB/N011600/1 to IHM], and the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001003), the UK Medical Research Council (FC001003), and the Wellcome Trust (FC001003) [R.A.G.C., M.T. and P.A.B.].
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Moal, I.H., Chaleil, R.A.G., Torchala, M., Bates, P.A. (2020). A Guide for Protein–Protein Docking Using SwarmDock. In: Kihara, D. (eds) Protein Structure Prediction. Methods in Molecular Biology, vol 2165. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0708-4_11
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0708-4_11
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0707-7
Online ISBN: 978-1-0716-0708-4
eBook Packages: Springer Protocols