Abstract
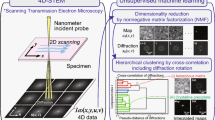
Scanning precession electron diffraction involves the acquisition of a two-dimensional precession electron diffraction pattern at every probe position in a two-dimensional scan. The data typically comprise many more diffraction patterns than the number of distinct microstructural volume elements (e.g. crystals) in the region sampled. A dimensionality reduction, ideally to one representative diffraction pattern per distinct element, may then be sought. Further, some diffraction patterns will contain contributions from multiple crystals sampled along the beam path, which may be unmixed by harnessing this oversampling. Here, we report on the application of unsupervised machine learning methods to achieve both dimensionality reduction and signal unmixing. Potential artefacts are discussed and precession electron diffraction is demonstrated to improve results by reducing the impact of bending and dynamical diffraction so that the data better approximate the case in which each crystal yields a given diffraction pattern.
Similar content being viewed by others
Introduction
Scanning transmission electron microscopy (STEM) investigations increasingly combine the measurement of multiple analytical signals as a function of probe position with post-facto computational analysis [1]. In a scan, the number of local signal measurements is usually much greater than the number of significantly distinct microstructural elements and this redundancy may be harnessed during analysis, for example by averaging signals over like regions to improve signal to noise. Unsupervised machine learning techniques automatically exploit data redundancy to find patterns with minimal prior constraints [2]. In analytical electron microscopy, such methods have been applied to learn representative signals corresponding to separate microstructural elements (e.g. crystal phases) and to unmix signals comprising contributions from multiple microstructural elements sampled along the beam path [3,4,5,6,7,8,9,10]. These studies have primarily applied linear matrix decompositions such as independent component analysis (ICA) and non-negative matrix factorisation (NMF).
Scanning precession electron diffraction (SPED) enables nanoscale investigation of local crystallography [11, 12] by recording electron diffraction patterns as the electron beam is scanned across the sample with a step size on the order of nanometres. The incorporation of double conical rocking of the beam, also known as precession [13], achieves integration through a reciprocal space volume for each reflection. Precession has been found to convey a number of advantages for interpretation and analysis of the resultant diffraction patterns, in particular the suppression of intensity variation due to dynamical scattering [14,15,16]. The resultant four-dimensional dataset, comprising two real and two reciprocal dimensions (4D-SPED), can be analysed in numerous ways. For example, the intensity of a sub-set of pixels in each diffraction pattern can be integrated (or summed) as a function of probe position, to form so-called virtual bright field (VBF) or virtual dark field (VDF) images [17, 18]. VBF/VDF analysis has been used to provide insight into local crystallographic variations such as phase [17], strain [19] and orientation [20]. In another approach, the collected diffraction patterns are compared against a library of precomputed templates, providing a visualisation of the microstructure and orientation information, a process known as template or pattern matching [11]. These analyses do not utilise the aforementioned redundancy present in data and may require significant effort on the part of the researcher. Here, we explore the application of unsupervised machine learning methods to achieve dimensionality reduction and signal unmixing.
Methods
Materials
GaAs (cubic, F43m) nanowires containing type I twin (\(\Sigma 3\)) [21] boundaries were taken as a model system for this work. The long axis of these nanowires is approximately parallel to the [111] crystallographic direction as a result of growth by molecular beam epitaxy [22] on (111). In cross section, these nanowires have an approximately hexagonal geometry with a vertex-to-vertex distance of 120–150 nm. Viewed near to the \([1{\overline{1}}0]\) zone axis, the twin boundary normal is approximately perpendicular to the incident beam direction.
SPED experiments
Scanning precession electron diffraction was performed on a Philips CM300 FEGTEM operating at 300 kV with the scan and simultaneous double rocking of the electron beam controlled using a NanoMegas Digistar external scan generator. A convergent probe with convergence semi-angle of \(\sim 1.5\hbox { mrad}\) and precession angles of 0, 9 and 35 mrad was used to perform scans with a step size of 10 nm using the ASTAR software package. The resolution was thus dominated by the step size. PED patterns were recorded using a Stingray CCD camera to capture the image on the fluorescent binocular viewing screen.
It is generally inappropriate to manipulate raw data before applying multivariate methods such as decomposition or clustering, which cannot be considered objective if subjective prior alterations have been made. In this work, the only data manipulation applied before machine learning is to align the central beams of each diffraction pattern. Geometric distortions introduced from the angle between the camera and the viewing screen were corrected by applying an opposite distortion to the data after the application of machine learning methods.
Multislice simulations
A twinned bi-crystal model was constructed with the normal to the [111] twin boundary inclined at an angle of \(55 ^{\circ }\) to the incident beam direction so that the two crystals overlapped in projection. In this geometry, both crystals are oriented close to \(\langle 511\rangle\) zone axes with coherent matching of the \(\{0{\bar{6}}6\}\) and \(\{2{\bar{8}}2\}\) planes in these zones. Three precession angles were simulated using the TurboSlice package [23]: 0, 10 and 20 mrad, with 200 distinct azimuthal positions about the optic axis to ensure appropriate integration in the resultant simulated patterns [24]. The crystal model used in the simulation comprised 9 unique layers each 0.404 nm thick. 15 layers were used leading to a total thickness of 54.6 nm. These \(512\times 512\)-pixel patterns with 16-bit dynamic range were convolved with a 4-pixel Gaussian kernel to approximate a point spread function.
Linear matrix decomposition
Latent linear models describe data by the linear combination of latent variables that are learned from the data rather than measured—more pragmatically, the repeated features in the data can be well approximated using a small number of basis vectors. With appropriate constraints, the basis vectors may be interpreted as physical signals. To achieve this, a data matrix, \(\mathbf X\), can be approximated as the matrix product of a matrix of basis vectors \(\mathbf W\) (components), and corresponding coefficients \(\mathbf Z\) (loadings). The error in the approximation, or reconstruction error, may be expressed as an objective function to be minimised in a least squares scheme:
where \(||\mathbf A ||_{\mathrm {F}}\) is the Frobenius normFootnote 1 of matrix \(\mathbf A\). More complex objective functions, for example incorporating sparsity promoting weighting factors [25], may be defined. We note that the decomposition is not necessarily performed by directly computing this error minimisation.
Three linear decompositions were used here: singular value decomposition (SVD) [2, 26], independent component analysis (ICA) [27], and non-negative matrix factorisation (NMF) [25, 28]. These decompositions were used as implemented in HyperSpy [29], which itself draws on the algorithms implemented in the open-source package scikit-learn [30].
The singular value decomposition is closely related to the better-known principal component analysis, in which the vectors comprising \(\mathbf W\) are orthonormal. The optimal solution to rank L is then obtained when \(\mathbf W\) is estimated by eigenvectors (principal components) corresponding to the L largest eigenvalues of the empirical covariance matrixFootnote 2. The optimal low-dimensional representation of the data is given by \(\mathbf z _{i} = \mathbf W ^{T}{} \mathbf x _{i}\), which is an orthogonal projection of the data on to the corresponding subspace and maximises the statistical variance of the projected data. This optimal reconstruction may be obtained via truncated SVD of the data matrix—the factors for PCA and SVD are equivalent, though the loadings may differ by independent scaling factors [\([1{\overline{1}}0]\) zone axis by \(\sim 30 ^{\circ }\), such that two microstructural elements overlapped in projection. The overlap of the two crystals was assessed using virtual dark-field imaging, NMF loading maps, and fuzzy clustering membership maps (Fig. 4). The region in which the crystals overlap can be identified by all these methods. The VDF result can be considered a reference and is obtained with minimal processing but requires manual specification of appropriate diffracting conditions for image formation. The NMF and fuzzy clustering approaches are semi-automatic. There is good agreement between the VDF images and NMF loading maps. The boundary appears slightly narrower in the clustering membership map. The NMF loading corresponding to the background component decreases along the profile, which may be related to the underlying carbon film, whilst the cluster membership for the background contains a spurious peak in the overlap region. Finally, the direct beam intensity is much lower in the NMF component patterns than in the true source signals. Our results indicate that either machine learning method is superior to conventional linear decomposition for the analysis of SPED datasets, but some unintuitive and potentially misleading features are present in the learning results.
SPED data from a GaAs nanowire with a twin boundary at an oblique angle to the beam. a Virtual dark-field images formed, using a virtual aperture 4 pixels in diameter, from the circled diffraction spots. b NMF decomposition results. c Clustering results. For b and c the profiles are taken from the line scans indicated, and the blue profile represents the intensity of the background component. Pattern and loading scale bars are common to all subfigures and measure 1 Å\(^{-1}\) and 70 nm respectively
Discussion
Unsupervised learning methods (SVD, ICA, NMF, and fuzzy clustering) have been explored here in application to SPED data as applied to materials where the region of interest comprises a finite number of significantly different microstructrual elements, i.e. crystals of particular phase and/or orientation. In this case, NMF and clustering may yield a small number of component patterns and weighted average cluster centres that resemble physical electron diffraction patterns. These methods are, therefore, effective for both dimensionality reduction and signal unmixing although we note that neither approach is well suited to situations where there are continuous changes in crystal structure. By contrast, SVD and ICA provide effective means of dimensionality reduction but the components are not readily interpreted using analogous methods to conventional electron diffraction analysis, owing to the presence of many negative values. The SVD and ICA results do nevertheless tend to highlight physically important differences in the diffraction signal across the region of interest. The massive data reduction from many thousands of measured diffraction patterns to a handful of learned component patterns is very useful, as is the unmixing achieved. Artefacts in the learning results were however identified, particularly when applied to achieve signal unmixing, and these are explored further here.
To illustrate artefacts resulting from learning methods, model SPED datasets were constructed based on line scans across inclined boundaries in hypothetical bicrystals. Models (Figs. 5 and 6) were designed to highlight features of two-dimensional diffraction-like signals rather than to reflect the physics of diffraction. These were, therefore, constructed with the strength of the signal directly proportional to thickness of the hypothetical crystal at each point, with no noise, and Gaussian peak profiles.
Construction and decomposition of an idealised model SPED dataset system comprising non-overlap** two-dimensional signals. a Schematic representation of hypothetical bi-crystal. b Ground truth end-member patterns and relative thickness of the two crystals. c Factors and loadings obtained by 2-component NMF. d Cluster centre average patterns and membership maps obtained by fuzzy clustering
Non-independent components. a Expected result for an artificial dataset with two ‘phases’ with overlap** peaks. b NMF decomposition. c Cluster results. d SVD loadings of the dataset, used for clustering. Each point corresponds to a diffraction pattern in the scan—several are indicated with the dotted lines. Contours indicate the value of membership to the two clusters—refer to “Methods” section “Data clustering”
The model SPED dataset shown in Fig. 5 comprises the linear summation of two square arrays of Gaussians (to emulate diffraction patterns) with no overlap between the two patterns. NMF decomposition exactly recovers the signal profile in this simple case. In contrast, the membership profile obtained by fuzzy clustering, which varies smoothly owing to the use of a Euclidean distance metric, does not match the source signal. The boundary region instead appears qualitatively narrower than the true boundary. Further, the membership value for each of the pure phases is slightly below 100% because the cluster centre is a weighted average position that will only correspond to the end member if there are many more measurements near to it than away from it. A related effect is that the membership value rises at the edge of the boundary region where mixed patterns are closer to the weighted centre than the end members. We conclude that clustering should be used only if the data comprises a significant amount of unmixed signal. In the extreme, cluster analysis cannot separate the contribution from a microstructural feature which has no pure signal in the scan, for example a fully embedded particle. These observations are consistent with the results reported in association with Fig. 4.
A common challenge for signal separation arises when the source signals contain coincident peaks from distinct microstructural elements, as would be the case in SPED data when crystallographic orientation relationships exist between crystals. A model SPED dataset corresponding to this case was constructed and decomposed using NMF and fuzzy clustering (Fig. 6). In this case, the NMF decomposition yields a factor containing all the common reflections and a factor containing the reflections unique to only one end member. Whilst this is interpretable, it is not physical, although it should be noted that this is an extreme example where there is no unique information in one of the source patterns. Nevertheless, it should be expected that the intensity of shared peaks is likely to be unreliable in the learned factors and this was the case for the direct beam in learned component patterns shown in Fig. 4. As a result, components learned through NMF should not be analysed quantitativelyFootnote 3. The weighted average cluster centres resemble the true end members much more closely than the NMF components. The pure phases have a membership of around 99%, rather than 100%, due to the cluster centre being offset from the pure cluster by the mixed data, as shown in Fig. 6d. The observation that memberships extend across all the data (albeit sometimes with vanishingly small values) explains the rise in intensity of the background component in Fig. 4c in the interface region. Such interface regions do not evenly split their membership between their two true constituent clusters, meaning that some membership is attributed to the third cluster, causing a small increase in the membership locally. These issues may potentially be addressed using extensions to the algorithm developed by Rousseeuw et al. [41] or using alternative geometric decompositions such as vertex component analysis [42].
Precession was found empirically to improve machine learning decomposition as discussed above (Fig. 2), so long as the precession angle is large enough. This was attributed primarily to integration through bending of the nanowire. Precession may also result in a more monotonic variation of diffracted intensity with thickness [15] as a result of integration through the Bragg condition. It was, therefore, suggested that precession may improve the approximation that signals from two overlap** crystals may be considered to be combined linearly. To explore this, a multislice simulation of a line scan across a bi-crystal was performed and decomposed using both NMF and fuzzy clustering (Fig. 7). Without precession, both the NMF loadings and the cluster memberships do not increase monotonically with thickness but rather vary significantly in a manner reminiscent of diffracted intensity modulation with thickness due to dynamical scattering. Both the loading profile and the membership profile reach subsidiary minima when the corresponding component is just thicker than half the thickness of the simulation, which corresponds to a thickness of approximately 100 nm and is consistent with the \(2{\bar{2}}0\) extinction length for GaAs of 114 nm. This suggests that the decomposition of the diffraction patterns is highly influenced by a few strong reflections; hence, the variation of the \(2{\bar{2}}0\) reflections with thickness is overwhelming the other structural information encoded in the patterns. The removal of this effect, an essential function of applying precession, is seen: with 10 or 20 mrad precession this intensity modulation is suppressed and the loading or membership maps obtained show a monotonic increase across the inclined boundary. The cluster centres again show intensity corresponding to the opposite end member due to the weighted averaging. Precession is, therefore, beneficial for the application of unsupervised learning algorithms both in reducing signal variations arising from bending, which is a common artefact of specimen preparation, and reducing the impact of dynamical effects on signal mixing.
Unsupervised learning applied to SPED data simulated using dynamical multislice calculations a Original data with a 20 mrad precession angle. b NMF decomposition, in which the loadings have been re-scaled as in Fig. 5. The factors show pseudo-subtractive features, typical of NMF. c Cluster analysis. The high proportion of data points from the boundary means there is information shared between the cluster centres. Without precession, neither method can reproduce the original data structure
Noise and background are both significant in determining the performance of unsupervised learning algorithms. Extensive exploration of these parameters is beyond the scope of this work but we note that the various direct electron detectors that have recently been developed and that are likely to play a significant role in future SPED studies have very different noise properties. Therefore, understanding the optimal noise performance for unsupervised learning may become an important consideration. We also note that the pseudo-subtractive features evident in the NMF decomposition results of Fig. 3 may become more significant in this case and the robustness of fuzzy clustering to this may prove advantageous.
Conclusions
Unsupervised machine learning methods, particularly non-negative matrix factorisation and fuzzy clustering, have been demonstrated here to be capable of learning the significant microstructural features within SPED data. NMF may be considered a true linear unmixing whereas fuzzy clustering, when applied to learn representative patterns, is essentially an automated way of performing a weighted averaging with the weighting learned from the data. The former can struggle to separate coincident signals (including signal shared with a background or noise) whereas the latter implicitly leaves some mixing when a large fraction of measurements are mixed. In both cases, precession electron diffraction patterns are more amenable to unsupervised learning than the static beam equivalents. This is due to the integration through the Bragg condition, resulting from rocking the beam, causing diffracted beam intensities to vary more monotonically with thickness and the integration through small orientation changes due to out of plane bending. This work has, therefore, demonstrated that unsupervised machine learning methods, when applied to SPED data, are capable of reducing the data to the most salient structural features and unmixing signals. The scope for machine learning to reveal nanoscale crystallography will expand rapidly in the coming years with the application of more advanced methods.
Notes
The Frobenius norm is defined as \(||\mathbf A ||_{\mathrm {F}} = \sqrt{\sum \limits _{i=1}^{m} \sum \limits _{j=1}^{n} a_{ij}^{2}}\)
The empirical covariance matrix, \(\Sigma = \frac{1}{N}\sum _{i=1}^{N} \mathbf x _{i}{} \mathbf x _{i}^{T}\)
This problem may be mitigated by enforcing a sum-to-one constraint on the loadings learned through NMF during optimisation. See for example [40].
Abbreviations
- BSS:
-
blind source separation
- FEGTEM:
-
field-emission gun transmission electron microscope
- ICA:
-
independent component analysis
- NNDSVD:
-
non-negative double singular value decomposition
- NMF:
-
non-negative matrix factorisation
- PED:
-
precession electron diffraction
- PCA:
-
principal component analysis
- SPED:
-
scanning precession electron diffraction
- STEM:
-
scanning transmission electron microscopy
- SVD:
-
singular value decomposition
- VBF:
-
virtual bright field
- VDF:
-
virtual dark field
References
Thomas, J.M., Leary, R.K., Eggeman, A.S., Midgley, P.A.: The rapidly changing face of electron microscopy. Chem. Phys. Lett. 631, 103–113 (2015). https://doi.org/10.1016/j.cplett.2015.04.048
Murphy, K.P.: Machine Learning: A Probabilistic Perspective. Adaptive Computation and Machine Learning. MIT Press, Boston (2012)
de la Peña, F., Berger, M.H., Hochepied, J.F., Dynys, F., Stephan, O., Walls, M.: Map** titanium and tin oxide phases using EELS: an application of independent component analysis. Ultramicroscopy 111(2), 169–176 (2011). https://doi.org/10.1016/J.ULTRAMIC.2010.10.001
Nicoletti, O., de la Peña, F., Leary, R.K., Holland, D.J., Ducati, C., Midgley, P.A.: Three-dimensional imaging of localized surface plasmon resonances of metal nanoparticles. Nature 502(7469), 80–84 (2013). https://doi.org/10.1038/nature12469
Rossouw, D., Burdet, P., de la Peña, F., Ducati, C., Knappett, B.R., Wheatley, A.E.H., Midgley, P.A.: Multicomponent signal unmixing from nanoheterostructures: overcoming the traditional challenges of nanoscale X-ray analysis via machine learning. Nano Lett. 15(4), 2716–2720 (2015). https://doi.org/10.1021/acs.nanolett.5b00449
Rossouw, D., Krakow, R., Saghi, Z., Yeoh, C.S., Burdet, P., Leary, R.K., de la Peña, F., Ducati, C., Rae, C.M., Midgley, P.A.: Blind source separation aided characterization of the \(\gamma\)’ strengthening phase in an advanced nickel-based superalloy by spectroscopic 4D electron microscopy. Acta Mater. 107, 229–238 (2016). https://doi.org/10.1016/j.actamat.2016.01.042
Rossouw, D., Knappett, B.R., Wheatley, A.E.H., Midgley, P.A.: A new method for determining the composition of core-shell nanoparticles via dual-EDX+EELS spectrum imaging. Particle Particle Syst. Charact. 33(10), 749–755 (2016). https://doi.org/10.1002/ppsc.201600096
Shiga, M., Tatsumi, K., Muto, S., Tsuda, K., Yamamoto, Y., Mori, T., Tanji, T.: Sparse modeling of EELS and EDX spectral imaging data by nonnegative matrix factorization. Ultramicroscopy 170, 43–59 (2016). https://doi.org/10.1016/J.ULTRAMIC.2016.08.006
Eggeman, A.S., Krakow, R., Midgley, Pa: Scanning precession electron tomography for three-dimensional nanoscale orientation imaging and crystallographic analysis. Nat. Commun. 6, 7267 (2015). https://doi.org/10.1038/ncomms8267
Sunde, J.K., Marioara, C.D., Van Helvoort, A.T.J., Holmestad, R.: The evolution of precipitate crystal structures in an Al-Mg-Si(-Cu) alloy studied by a combined HAADF-STEM and SPED approach. Mater. Charact. 142, 458–469 (2018). https://doi.org/10.1016/j.matchar.2018.05.031
Rauch, E.F., Veron, M.: Coupled microstructural observations and local texture measurements with an automated crystallographic orientation map** tool attached to a TEM. Materialwissenschaft und Werkstofftechnik 36(10), 552–556 (2005). https://doi.org/10.1002/mawe.200500923
Rauch, E.F., Portillo, J., Nicolopoulos, S., Bultreys, D., Rouvimov, S., Moeck, P.: Automated nanocrystal orientation and phase map** in the transmission electron microscope on the basis of precession electron diffraction. Zeitschrift für Kristallographie 225(2–3), 103–109 (2010). https://doi.org/10.1524/zkri.2010.1205
Vincent, R., Midgley, P.: Double conical beam-rocking system for measurement of integrated electron diffraction intensities. Ultramicroscopy 53(3), 271–282 (1994). https://doi.org/10.1016/0304-3991(94)90039-6
White, T., Eggeman, A., Midgley, P.: Is precession electron diffraction kinematical? Part I: “Phase-scrambling” multislice simulations. Ultramicroscopy 110(7), 763–770 (2010). https://doi.org/10.1016/J.ULTRAMIC.2009.10.013
Eggeman, A.S., White, T.A., Midgley, P.A.: Is precession electron diffraction kinematical? Part II. A practical method to determine the optimum precession angle. Ultramicroscopy 110(7), 771–777 (2010). https://doi.org/10.1016/j.ultramic.2009.10.012
Sinkler, W., Marks, L.D.: Characteristics of precession electron diffraction intensities from dynamical simulations. Zeitschrift für Kristallographie 225(2–3), 47–55 (2010). https://doi.org/10.1524/zkri.2010.1199
Rauch, E.F., Véron, M.: Virtual dark-field images reconstructed from electron diffraction patterns. Eur. Phys. J. Appl. Phys. 66(1), 10,701 (2014). https://doi.org/10.1051/epjap/2014130556
Gammer, C., Burak Ozdol, V., Liebscher, C.H., Minor, A.M.: Diffraction contrast imaging using virtual apertures. Ultramicroscopy 155, 1–10 (2015). https://doi.org/10.1016/J.ULTRAMIC.2015.03.015
Rouviere, J.L., Béché, A., Martin, Y., Denneulin, T., Cooper, D.: Improved strain precision with high spatial resolution using nanobeam precession electron diffraction. Appl. Phys. Lett. 103(24), 241913 (2013). https://doi.org/10.1063/1.4829154
Moeck, P., Rouvimov, S., Rauch, E.F., Véron, M., Kirmse, H., Häusler, I., Neumann, W., Bultreys, D., Maniette, Y., Nicolopoulos, S.: High spatial resolution semi-automatic crystallite orientation and phase map** of nanocrystals in transmission electron microscopes. Crys. Res. Technol. 46(6), 589–606 (2011). https://doi.org/10.1002/crat.201000676
Kelly, A., Groves, G., Kidd, P.: Crystallography and Crystal Defects. Wiley, Chichester (2000)
Munshi, A.M., Dheeraj, D.L., Fauske, V.T., Kim, D.C., Huh, J., Reinertsen, J.F., Ahtapodov, L., Lee, K.D., Heidari, B., van Helvoort, A.T.J., Fimland, B.O., Weman, H.: Position-controlled uniform GaAs nanowires on silicon using nanoimprint lithography. Nano Lett. 14(2), 960–966 (2014). https://doi.org/10.1021/nl404376m
Eggeman, A., London, A., Midgley, P.: Ultrafast electron diffraction pattern simulations using gpu technology. Applications to lattice vibrations. Ultramicroscopy 134, 44–47 (2013). https://doi.org/10.1016/j.ultramic.2013.05.013
Palatinus, L., Jacob, D., Cuvillier, P., Klementová, M., Sinkler, W., Marks, L.D.: IUCr: structure refinement from precession electron diffraction data. Acta Crystallogr. Sect. A Found. Crystallogr. 69(2), 171–188 (2013). https://doi.org/10.1107/S010876731204946X
Hoyer, P.O.: Non-negative matrix factorization with sparseness constraints. J. Mach. Learn. Res. 5, 1457–1469 (2004)
Jolliffe, I.: Principal component analysis. In: International Encyclopedia of Statistical Science, pp. 1094–1096. Springer , Berlin (2011)
Hyvärinen, A., Karhunen, J., Oja, E.: Independent Component Analysis. Wiley, New York (2001)
Lee, D.D., Seung, H.S.: Learning the parts of objects by non-negative matrix factorization. Nature 401(6755), 788–91 (1999). https://doi.org/10.1038/44565
de la Pena, F., Ostasevicius, T., Tonaas Fauske, V., Burdet, P., Jokubauskas, P., Nord, M., Sarahan, M., Prestat, E., Johnstone, D.N., Taillon, J., Jan Caron, J., Furnival, T., MacArthur, K.E., Eljarrat, A., Mazzucco, S., Migunov, V., Aarholt, T., Walls, M., Winkler, F., Donval, G., Martineau, B., Garmannslund, A., Zagonel, L.F., Iyengar, I.: Electron Microscopy (Big and Small) Data Analysis With the Open Source Software Package HyperSpy. Microsc. Microanal. 23(S1), 214–215 (2017). https://doi.org/10.1017/S1431927617001751
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., Vanderplas, J., Passos, A., Cournapeau, D., Brucher, M., Perrot, M., Duchesnay, É.: Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12(Oct), 2825–2830 (2011)
Shlens, J.: A tutorial on principal component analysis. CoRR (2014). ar**v:abs/1404.1100
Bishop, C.: Pattern Recognition and Machine Learning. Information Science and Statistics. Springer, New York (2006)
Boutsidis, C., Gallopoulos, E.: Svd based initialization: a head start for nonnegative matrix factorization. Pattern Recogn. 41(4), 1350–1362 (2008). https://doi.org/10.1016/j.patcog.2007.09.010
Everitt, B., Landau, S., Leese, M.: Clust. Anal. Wiley, Chichester (2009)
Bezdek, J.C., Ehrlich, R., Full, W.: FCM: the fuzzy c-means clustering algorithm. Comput. Geosci. 10(2–3), 191–203 (1984). https://doi.org/10.1016/0098-3004(84)90020-7
Gustafson, D., Kessel, W.: Fuzzy clustering with a fuzzy covariance matrix. In: 1978 IEEE conference on decision and control including the 17th symposium on adaptive processes, pp. 761–766. IEEE, San Diego (1978). https://doi.org/10.1109/CDC.1978.268028
Marimont, R.B., Shapiro, M.B.: Nearest neighbour searches and the curse of dimensionality. IMA J. Appl. Math. 24(1), 59–70 (1979). https://doi.org/10.1093/imamat/24.1.59
Aggarwal, C.C., Hinneburg, A., Keim, D.A.: On the surprising behavior of distance metric in high-dimensional space (2002)
Rencher, A.: Methods of Multivariate Analysis. Wiley Series in Probability and Statistics. Wiley, Hoboken (2002)
Kannan, R., Ievlev, A.V., Laanait, N., Ziatdinov, M.A., Vasudevan, R.K., Jesse, S., Kalinin, S.V.: Deep data analysis via physically constrained linear unmixing: universal framework, domain examples, and a community-wide platform. Adv. Struct. Chem. Imaging 4(1), 6 (2018). https://doi.org/10.1186/s40679-018-0055-8
Rousseeuw, P.J., Trauwaertb, E., Kaufman, L.: Fuzzy clustering with high contrast. J. Comput. Appl. Math. 0427(95), 8–9 (1995)
Spiegelberg, J., Rusz, J., Thersleff, T., Pelckmans, K.: Analysis of electron energy loss spectroscopy data using geometric extraction methods. Ultramicroscopy 174, 14–26 (2017). https://doi.org/10.1016/J.ULTRAMIC.2016.12.014
Authors' contributions
BHM, DNJ, ASE, and PAM proposed the investigation. DNJ, ASE, and ATJvH performed the SPED experiments, ASE provided the multislice simulations, and BHM implemented the c-means algorithm. The data analysis was undertaken by BHM and DNJ who also prepared the manuscript, with oversight and critical contributions from ASE, ATJvH, and PAM. All authors read and approved the final manuscript.
Acknowlegements
Prof. Weman and Fimland of IES at NTNU are acknowledged for supplying the nanowire samples.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data used in this work has been made freely available to download at https://doi.org/10.17863/CAM.26432.
The Python 3 code to perform the analysis has also been made available, at https://doi.org/10.17863/CAM.26444.
Funding
The authors acknowledge financial support from: The Royal Society (Grant RG140453; UF130286); the Seventh Framework Programme of the European Commission: ESTEEM2, contract number 312483; the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement 291522-3DIMAGE; the University of Cambridge and the Cambridge NanoDTC; the EPSRC (Grant no. EP/R008779/1).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Martineau, B.H., Johnstone, D.N., van Helvoort, A.T.J. et al. Unsupervised machine learning applied to scanning precession electron diffraction data. Adv Struct Chem Imag 5, 3 (2019). https://doi.org/10.1186/s40679-019-0063-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40679-019-0063-3