Abstract
Background
To evaluate the performance of diffusion-relaxation correlation spectrum imaging (DR-CSI) with support vector machine (SVM) in detecting prostate cancer (PCa).
Methods
In total, 114 patients (mean age, 66 years, range, 48–87 years) who received a prostate MRI and underwent biopsy were enrolled in three stages. Thirty-nine were assigned for the exploration stage to establish the model, 18 for the validation stage to choose the appropriate scale for map** and 57 for the test stage to compare the diagnostic performance of the DR-CSI and PI-RADS.
Results
In the exploration stage, the DR-CSI model was established and performed better than the ADC and T2 values (both P < 0.001). The validation result shows that at least 2 pixels were required for both the long-axis and short-axis in the map** procedure. In the test stage, DR-CSI had higher accuracy than PI-RADS ≥ 3 as a positive finding based on patient (84.2% vs. 63.2%, P = 0.004) and lesion (78.8% vs. 57.6%, P = 0.001) as well as PI-RADS ≥ 4 on lesion (76.5% vs. 64.7%, P = 0.029), while there was no significant difference between DR-CSI and PI-RADS ≥ 4 based on patient (P = 0.508). For clinically significant PCa, DR-CSI had higher accuracy than PI-RADS ≥ 3 based on patients (84.2% vs. 63.2%, P = 0.004) and lesions (62.4% vs. 48.2%, P = 0.036). There was no significant difference between DR-CSI and PI-RADS ≥ 4 (P = 1.000 and 0.845 for the patient and lesion levels, respectively).
Conclusions
DR-CSI combined with the SVM model may improve the diagnostic accuracy of PCa.
Trial registration
This study was approved by the Ethics Committee of our institute (Approval No. KY2018-213). Written informed consent was obtained from all participants.
Similar content being viewed by others
Background
Prostate cancer (PCa) is the second most frequently occurring cancer among males, with the highest incidence rates in over 60% countries worldwide, and remains a leading cause of cancer-related death in many countries, imposing a heavy burden on families and communities [1]. The accurate detection of PCa at an earlier stage is of vital importance for clinical decision-making, the evaluation of long-term survival and better outcomes/prognosis. For clinical prostate examination, magnetic resonance imaging (MRI) is a significant imaging modality that provides superb soft tissue contrast and functional evaluation. To date, multiparametric MRI (MP-MRI), including T2-weighted imaging (T2WI), diffusion-weighted imaging (DWI), and dynamic contrast-enhanced MRI (DCE-MRI), has been proven to be a promising noninvasive tool for PCa detection [2,3,4].
Among the metrics derived from MP-MRI, T2 and apparent diffusion coefficient (ADC) are widely accepted as the two most valuable biomarkers for tumour characterization [4, 5]. Furthermore, both T2 and ADC have been found to be related to the aggressiveness of PCa, which is indicated by the Gleason score [6, 7]. However, with the increasing application of MP-MRI using the Prostate Imaging-Reporting and Data System Version 2.1 (PI-RADS v2.1) for PCa diagnosis, it was found that approximately 15–30% of clinically significant cancers were undetected, which, to some extent, depends on the clinical experience of the radiologists [8, 9]. As such, accurate PCa diagnosis remains a challenge [10,11,12].
Concerning PCa, multiple intravoxel components with different or similar MR properties, such as T2 or ADC, add difficulty for MP-MRI in differentiation. In addition, various tissue structural features at a microscopic scale produce another obstacle for visualization owing to limited MRI spatial resolution. To resolve this issue, some attempts have been made in multidimensional MRI, a method that tries to disentangle intravoxel signals in terms of parametric spectra, such as T1, T2 and ADC, and hence to infer the intravoxel tissue composition [13, 14]. For example, hybrid multidimensional MRI (HM-MRI) was used to probe the stroma, epithelium, and lumen changes in PCa [14, 15]. Moreover, the novel diffusion-relaxation correlation spectrum imaging (DR-CSI) method was proposed [16] and successfully identified and quantified multiple distinct components (epithelium, stroma, and lumen) in the ex vivo prostate through a T2–ADC spectrum with prior knowledge [17]. Owing to the heterogeneous tissue structure and biological environment in vivo, it is difficult to know the prior knowledge to make a tissue structural assumption about PCa based on which DR-CSI could apply. Nevertheless, to our knowledge, no research on DR-CSI for in vivo PCa detection has been initiated yet.
In this study, the machine learning technique, the support vector machine (SVM) model, was introduced in combination with DR-CSI. SVM, as a supervised ML technique that tries to find the optimal hyperplane that maximizes the margin between two classes, has been widely used for the detection and classification of PCa due to its mature mathematical formulation, flexibility, high accuracy, robust theoretical support, direct geometric interpretation, and wide availability of software implementations [18,19,20,21,22,23]. For example, integrating with the parameters of MP-MRI, such as diffusion and perfusion, SVM enabled accurate and automatic classification of low-grade and high-grade PCa in the central gland [23]. Combined with DR-CSI, the SVM model was used to analyse spectral results according to the PCa diagnosis results instead of direct biologic interpretation based on a tissue structural assumption. The purpose of this study was to evaluate the feasibility of DR-CSI combined with an SVM model for the detection of PCa in vivo and to compare its diagnostic performance with PI-RADS v2.1 scores.
Methods
Patients
This retrospective study was approved by the Ethics Committee of our institute (Approval No. KY2018-213). Written informed consent was obtained from all participants. Three hundred and twenty consecutive patients who were scheduled for prostate MRI were recruited for this study between August 2020 and December 2021. The inclusion criteria were as follows: (a) prostate-specific antigen (PSA) elevated to ≥ 4 ng/mL or digital rectal examination (DRE) positive; (b) complete MRI examination and clinical data; and (c) no surgery before MRI examination. The exclusion criteria were as follows: (a) images with poor quality or artefacts; (b) patients with a previous history of prostate biopsy, prostate surgery, or other treatment; and (c) time interval between MRI and biopsy ≥ 3 months. In this study, 12 patients with previous chemotherapy, 5 patients with previous operation or treatment for PCa, and 2 patients with poor quality or artefacts were excluded. Three hundred and one patients were included (100 for the exploration stage, 56 for the validation stage and 145 for the test stage). In total, 114 patients who underwent biopsy were enrolled in three stages. Detailed participant characteristics are summarized in Table 1.
Study design
After prostate MRI, patients were enrolled in three stages – the exploration, validation, and test stages. In the exploration stage, patients with PI-RADS ≥ 3 lesions or DRE positivity underwent biopsy, and a PCa detection model utilizing an SVM model was established based on the DR-CSI and biopsy results. Then, in the validation stage, an optimal filter scale for the SVM model was chosen with the images used to detect PCa from biopsy, in which the biopsies were carried out on patients with PI-RADS ≥ 3 lesions or positive results according to the PCa detection model or DRE positivity. Finally, in the test stage, the PCa detection model was used to predict PCa, and the results were compared with PI-RADS scores as well as the gold standard. The biopsy results of patients had the same criteria as in the validation stage. The comparison was performed based on patient level as well as lesion level. At the lesion level, a single lesion was defined as a positive targeted lesion in DR-CSI or PI-RADS. PCa proven in 12 systemic biopsies with negative imaging results was also defined as a single lesion. For patients biopsied due to positive DRE results with negative imaging and pathology results, the whole prostate was defined as a single case. The flowchart of the study design is presented in Fig. 1.
MR Imaging acquisition
All MRI examinations were performed on a 3.0 T scanner (uMR 780, United Imaging Healthcare, Shanghai, China) with a commercial body phased array coil. MR examination included T2WI, DWI, DCE-MRI, T1-weighted imaging (T1WI) and DR-CSI. T1WI and T2WI were acquired. The detailed MR protocols are presented in Table 2. DR-CSI was also performed based on the SS SE-EPI including a matrix of 6*6 data points of echo times (57, 75, 100, 120, 150, 180 ms) and b-values (0, 150, 400, 800, 1200, 1500 s/mm2). The SS SE-EPI sequence was performed 6 times for each patient. For each time, TE was set as a specific value in its range with a full set of b-values (0, 150, 400, 800, 1200, 1500 s/mm2), i.e., diffusion sequence with different T2 weights. As TE was increased, diffusion time was increased in parallel.
DR-CSI Postprocessing
According to DR-CSI theory [16], the MR signal is expressed as an integral of exponential decay functions characterized by T2 and D:
where T2 and D represent the transverse relaxation time and the diffusivity, respectively. \(\mathrm{w}(\mathrm{x},\mathrm{y},\mathrm{T}2,\mathrm{D})\) stands for a 4D distribution function that connects the T2-D spectrum with each spatial location.
In practice, the signal needs to be expressed in a discretized form:
where I and J are the numbers of T2 and D, respectively. T2i and Dj represent the transverse relaxivity and diffusivity, respectively. \({\mathrm{w}}_{\mathrm{i},\mathrm{j}}(\mathrm{x},\mathrm{y},\mathrm{T}{2}_{\mathrm{i}},{\mathrm{D}}_{\mathrm{j}})\) is a discrete 4D distribution function described above.
The distribution function was obtained through spectrum estimation, i.e., finding the solution to the above equation with consistency, a nonnegativity and a spatial smoothness constraint following steps in a previous study [16].
To analyse component distributions in regions of interest (ROIs), the T2-D spectra space was empirically segmented into six subregions (i.e., components). A previous study demonstrated that the epithelial structure was more likely to be present within the scale of T2 < 50 ms and D < 0.5 mm2/μs based on ex vivo imaging [18]. Considering the enrichment of epithelial structure in PCa [14, 15], these two values were selected as the cut-offs for further classification. Moreover, D = 3 mm2/μs was also adopted as the cut-off value because the diffusion velocity of water molecules beyond 3 mm2/μs may likely characterize particular physiological information (e.g., perfusion, secretion). Thus, the ranges of the subregions were finally classified as follows: 1) T2 < 50 ms, D < 0.5 mm2/μs; 2) T2 > 50 ms, D < 0.5 mm2/μs; 3) T2 < 50 ms, 0.5 mm2/μs < D < 3 mm2/μs; 4) T2 > 50 ms, 0.5 mm2/μs < D < 3 mm2/μs; 5) T2 < 50 ms, D > 3 mm2/μs; and 6) T2 > 50 ms, D > 3 mm2/μs. The corresponding fraction of each subregion, (\({f}_{i}, i=\mathrm{1,2},\dots 6\)), for each voxel was estimated by the normalization of distribution functions:
where \(i\) denotes the order of the segmented subregion of the T2-D spectral space.
Image analysis
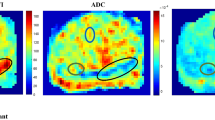
Figure 2 presents the whole process of image analysis. All image analyses were processed using MATLAB R2021a software (MathWorks, Natick, MA, USA). In the exploration and validation stage, the saturation prostate biopsy scheme was performed with US-MRI fusion biopsy (Esaote, Genoa, Italy). The median number of biopsy cores was 23. The location of the biopsy was further remarked on MRI [24]. Then, an experienced radiologist (G. W with 14 years of experience in prostate MR imaging) drew the ROI presented with PI-RADs ≥ 3 within 3 mm from the positive core of biopsies based on DW image with b-value = 1500 s/mm2 with the lowest TE (57 ms) [25,26,27]. For the cases with a negative biopsy result, circular ROIs with a radius from 3 to 8 mm were delineated on the bilateral central and peripheral areas in the largest area of the prostate (Fig. 3). Patients in the test stage underwent 12 core systemic biopsies, combined with targeted biopsies from MP-MRI and DR-CSI, which we designed to assess, with each core identified and processed separately. The ROIs of the whole prostate were delineated in the test stages. ROI drawn by another radiologist (with 11 years of experience in prostate MR imaging) was used to assess the interobserver agreement of the technique.
Accordingly, the fraction maps corresponding to six subregions of each ROI were acquired through DR-CSI analysis. Meanwhile, conventional voxelwise T2 and ADC maps of all ROIs were estimated following a monoexponential signal decay model using part of the DR-CSI data points (b = 0, TE = 57/75/100/120/150/180 ms; TE = 57 ms, b = 0/800 s/mm2), respectively [7].
The SVM-related procedures were as follows. First, the quantitative imaging features (T2, ADC and six component fractions) of all voxels from all patients in the exploration stage were used for the construction of the PCa detection model. Then, an SVM classifier with a linear kernel for PCa prediction was trained and established through tenfold cross validation, which evenly split the dataset into 10 subsets with 9 subsets for training and the last subset for evaluation. This process was repeated 10 times, leaving one different subset for evaluation each time. Second, different filter scales with different numbers of pixels (i.e., 1*1, 1*2, 1*3, …, 5*5) were used in the validation stage to choose an optimal scale, which could reduce the false-positive results resulting from the SVM-based PCa detection model. Then, the optimal filter scale with the highest Dice ratio between the modelling results and biopsy results was determined. Third, the SVM-based PCa detection model with the optimal filter scale was used in the test stage, and the voxelwise model-predicted map** results were generated.
The PI-RADS score was determined by an experienced radiologist (with more than 10 years of experience in prostate MR imaging) unaware of the biopsy results in the exploration and validation stage. In the test stage, the GS of the lesions was assigned according to the most common GS in the biopsy result.
Statistical analysis
Statistical analysis was performed by SPSS 24.0 (IBM Corp, NY, USA). Kappa values < 0.20 indicated slight agreement, values of 0.21–0.40 indicated fair agreement, values of 0.41–0.60 indicated moderate agreement, values of 0.61–0.80 indicated substantial agreement, and values > 0.81 indicated almost perfect agreement [28]. In the exploration stage, the performance of DR-CSI was compared with traditional ADC and T2 maps in differentiating cancer from non-PCa tissue at the largest Youden index point in receiver operating characteristic (ROC) analysis based on each voxel. In the test stage, the comparison of accuracy between DR-CSI and PI-RADS in terms of the diagnosis of PCa or cs-PCa was assessed by the McNemar test. Differences were considered significant when the P value was < 0.05.
Results
In the exploration stage, a total of 39 patients underwent biopsy, among which 3 were PI-RADS 2 but DRE positive, 21 were PI-RADS 3, 10 were PI-RADS 4, and 5 were PI-RADS 5. There were 72 lesions (4 lesions with PI-RADS 2, 31 lesions with PI-RADS 3, 27 with PI-RADS 4, and 10 with PI-RADS 5). The maximal diameter of the index lesion ranged from 0.4–3.9 cm (median = 1.3 cm). Of these, 17 patients were confirmed to have benign disease (3 with PI-RADS 2, 10 with PI-RADS 3, 4 with PI-RADS 4), and 22 patients had PCa (11 with PI-RADS 3, 6 with PI-RADS 4, 5 with PI-RADS 5). There were 29 PCa lesions (12 lesions with PI-RADS 3, 9 with PI-RADS 4, and 8 with PI-RADS 5). A total of 10,434 tumour voxels (3453 with Gleason grade = 6, 3875 with Gleason grade ≥ 7, and 3106 non-PCa tissue voxels) were included in the analysis to establish an SVM model. Figure 4 shows the weights of T2 and ADC values and six subregions contributing to the SVM model. Figure 5 shows the diagnostic performance of the DR-CSI model, traditional ADC and T2 value. The DR-CSI model was more accurate than the traditional ADC (0.87 vs. 0.81, P < 0.001) and T2 value (0.87 vs. 0.70, P < 0.001) at the highest Youden index point. Interreader agreement analysis showed that almost perfect agreement was achieved in each subregion (kappa value = 0.99, 1.00, 0.98, 0.99, 0.99 and 0.98 for components 1–6, respectively).
In the validation stage, a total of 18 patients (17 patients with PI-RADS ≥ 3 or DR-CSI positivity and 1 patient with PI-RADS < 3 and DR-CSI negativity but DRE positivity) underwent biopsy. Of these, 6 patients were confirmed to have benign disease, and 12 had PCa. We chose those 12 images and compared the performance of different map** methods to choose an appropriate scale for further testing. Among each scale, the largest Dice ratio was found in the scale, with the long axis and the short axis having at least 2 and 2 pixels, respectively (Fig. 6). The scale was further applied in the test stage.
In the test stage, a total of 57 patients underwent biopsy, among which 7 patients were PI-RADS < 3 and DR-CSI negative but DRE positive, 1 was PI-RADS 2 but DR-CSI positive, 28 were PI-RADS 3, 13 were PI-RADS 4, and 8 were PI-RADS 5. The maximal diameter of the index lesion ranged from 0.3–3.2 cm with a median of 1.1 cm. Of these, 25 patients were confirmed to have benign disease (7 with PI-RADS 2, 11 with PI-RADS 3, 7 with PI-RADS 4), and 32 had PCa (1 with PI-RADS 2, 17 with PI-RADS 3, 6 with PI-RADS 4, 8 with PI-RADS 5). Eighty-five lesions were subsequently verified. Thirty-one were confirmed to be benign, and 54 were PCa. Two typical cases are represented in Fig. 7. We analysed separately based on patient and lesion (Table 3). Considering PI-RADS ≥ 3 as a positive finding, DR-CSI had higher accuracy than PI-RADS ≥ 3 based on patients (84.2% vs. 63.2%, P = 0.004) and lesions (78.8% vs. 57.6%, P = 0.001). Considering PI-RADS ≥ 4 as a positive finding, DR-CSI had higher accuracy than PI-RADS ≥ 4 based on lesions (76.5% vs. 64.7%, P = 0.029), while no significant difference was found based on patients (84.2% vs. 78.9%, P = 0.508). For the analysis of clinically significant PCa, considering PI-RADS ≥ 3 as a positive finding, DR-CSI had higher accuracy than PI-RADS ≥ 3 based on patients (84.2% vs. 63.2%, P = 0.004) and lesions (62.4% vs. 48.2%, P = 0.036). Considering PI-RADS ≥ 4 as a positive finding, there was no significant difference between DR-CSI and PI-RADS ≥ 4 based on patients (84.2% vs. 82.5%, P = 1.000) and lesions (62.5% vs. 64.7%, P = 0.845).
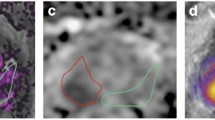
Examples of lesion detection. The first row shows a 74-year-old patient, tPSA 5.97 ng/mL, diagnosed with PI-RADS 3 at the right middle-anterior peripheral zone but negative in DR-CSI, who was finally confirmed to be benign by biopsy. The second row shows a 65-year-old patient, tPSA 7.79 ng/mL, diagnosed with PI-RADS 2 but positive in DR-CSI in the left middle peripheral zone, who was finally confirmed to have PCa and a Gleason score of 3 + 3 by biopsy. (a and f) T2WI; (b and g) diffusion weighted imaging; (c and h) apparent diffusion coefficient; (d and i) diffusion-relaxation correlation spectrum imaging (DR-CSI) result; (e and j) Decreasing of signal intensity in lesions with PI-RADs 3 delineated in the DR-CSI images (a & f)
Discussion
In this study, the feasibility of DR-CSI combined with an SVM model for detecting PCa in vivo was initially explored, and its diagnostic performance was evaluated and compared with the PI-RADS score based on MP-MRI. The optimal performance of the former method suggested additional clinical value and potential to improve the detection of PCa, especially based on lesion level. The results demonstrated that DR-CSI could provide additional information for PCa characterization in some clinical scenarios (e.g., targeted biopsy).
Conventionally, T2 and ADC derived from MP-MRI independently have been widely used for cancer detection and have proven to be of clinical significance [29, 30]. Some studies have reported that a strong interdependence exists between the measured ADC and T2 and varies depending on the tissue composition in PCa [31, 32]. Conventional MRI methods, which only provide voxel-averaged information on a macroscopic level, cannot disentangle intravoxel heterogeneity reflected by water diffusivity and relaxivity. In this sense, the ability of conventional MRI methods to investigate the tissue structure and composition in tumours is limited. In our study, compared with T2 and ADC maps alone, the diagnostic accuracy was improved by the DR-CSI with the SVM model. This could be attributed to the fact that DR-CSI could resolve the distribution of tissue components according to different spectral regions, providing additional information on tissue composition or structural features at a subvoxel level and hence improving the clinical diagnosis of PCa.
With decades of efforts, multidimensional MRI has been used to infer intravoxel heterogeneity in tissue due to its outstanding power to resolve signals from distinct tissue components. In pioneering work, DR-CSI has shown the potential to discriminate spatially overlap** phantoms and separate white and grey matter in both normal and injured animal spinal cords [16]. Specifically, with a priori knowledge, a hybrid multidimensional MRI model was set up based on an assumption of three different components in PCa [14, 15]. Owing to the intravoxel heterogeneity of tissue structure and the presence of multiple components in PCa (such as stroma, epithelium, and lumen, but not limited to these components) [33], the simplified DR-CSI model may limit the evaluation of microstructure complexity. Recently, some studies explored and validated the relations between the histological examination results and signal component fractions derived from DR-CSI in ex vivo PCa and in vivo PCa [15, 17].
However, the clinical diagnosis of PCa by DR-CSI still needs further study. Particularly, it is worth noting that there still exist other challenges to DR-CSI application for the clinical diagnosis of PCa [16]. First, the interpretation of the spectral results, i.e., associating the spectral peaks with specific tissue components or microstructure features, is not straightforward. Second, it is challenging to identify and determine the boundaries of spectral peaks, as the peaks would overlap and merge with one another, especially in malignant tumours with high intravoxel heterogeneity, making it difficult to robustly measure and map compartmental signal fractions.
Instead of assigning an individual spectral peak in the T2-D spectrum to a specific component, dividing the entire T2-D spectral space into six areas with different ranges of T2 and D could be a feasible way to reflect the diffusion-relaxation correlation information and avoid the need to identify specific components in the high intravoxel heterogeneous tissue structure. Thus, our DR-CSI was established based on segmented spectral areas to evaluate tissue composition in the diagnosis of PCa rather than to define a specific peak.
As the SVM model is an efficient way to deal with classification problems [22], it was adopted to build a voxel-based classification model for PCa diagnosis based on the resulting T2-D spectrum without defining and explaining the associations between segmented spectral areas with multiple components in PCa on the premise of no adequate prior knowledge about the microstructure compositions in vivo PCa. The results of the training model easily converged and could be quickly translated into practical applications. Moreover, the SVM model can explore the importance of a specific factor with the weight of each component in the model. ADC was still an important factor in PCa diagnosis, while different compartments derived from DR-CSI can simultaneously contribute to the classification model. Interestingly, components 5 and 6, which were considered to be associated with tissue perfusion in vivo with D > 3 mm2/μs, also accounted for significant proportions in the model. This finding verified the importance of perfusion emphasized by PI-RADS [10, 12]. Moreover, this result demonstrated that it is of great value to include the information available only in in vivo DR-CSI to improve the practical performance.
In the validation stage, we used 2*2 voxels as the filters to establish the map** model. It is important to choose an appropriate filter scale for image map** in SVM. Since the EPI sequence on the imaging level in vivo is easily disturbed by various artefacts, the direct map** method without adding filters may not reach satisfactory results, such as increased false-positive results. Lesions with 2*2 voxels will obviously provide more reliable information than lesions with a single voxel. The scale with 2*2 voxels is smaller than most prostate tumours that can be found by MRI and will not lead to missed diagnosis due to the choice of boundary.
In this study, we also evaluated the performance of the DR-CSI model in detecting cs-PCa. Although the performance of the DR-CSI model did not exceed that of PI-RADS, it still has some practical significance. As the result of the DR-CSI model can be automatically derived with SVM, the method also provides a potential idea to solve the problem of lacking reproducibility with PI-RADS in diagnosis [6]. Meanwhile, as the model was established to differentiate cancer lesions from benign lesions, the model certainly lacks high precision to distinguish between cc-PCa and cs-PCa. The performance in detecting cs-PCa may further improve if only lesions with GS ≥ 7 are used in modelling.
Our study had some limitations to be acknowledged. First, considering that the diagnostic performance of saturation biopsy was consistent with that of radical prostatectomy pathology in general [25, 34], saturation biopsy was adopted instead of whole-mount results in our study. However, the method can still omit some information on heterogeneity, the performance of the model might be improved with reference to the whole-mount result, and further study is needed to probe the issue. Second, although improved spectral sensitivity and resolution are expected from more data points, an increase in acquisition time cost is inevitable. Further research is needed to find the optimal choice of TE and b-values. Third, the impact of the DR-CSI protocol, for instance, the spatial resolution and SNR, on the spectral sensitivity and resolution of DR-CSI was not evaluated. Fourth, in this study, we adopt a method to divide the spectrum based on a priori knowledge, which is just one of many possible patterns. Further studies are needed to explore the optimal method of dividing the subareas of the spectrum. Investigation of the optimized DR-CSI protocol is warranted in future studies.
Conclusions
In conclusion, the DR-CSI combined with the SVM model has the potential to improve the diagnostic accuracy of prostate cancer.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DR-CSI:
-
Diffusion-relaxation correlation spectrum imaging
- SVM:
-
Support vector machine
- PCa:
-
Prostate cancer
- PI-RADS:
-
Prostate Imaging-Reporting and Data System
- MRI:
-
Magnetic resonance imaging
- MP-MRI:
-
Multiparametric MRI
- T2WI:
-
T2-weighted imaging
- T1WI:
-
T1-weighted imaging
- DWI:
-
Diffusion-weighted imaging
- DCE-MRI:
-
Dynamic contrast-enhanced MRI
- ADC:
-
Apparent diffusion coefficient
- HM-MRI:
-
Hybrid multidimensional MRI
- PSA:
-
Prostate-specific antigen
- DRE:
-
Digital rectal examination
- ROI:
-
Region of interest
- ROC:
-
Receiver operating characteristic
References
Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Caglic I, Barrett T. Optimising prostate mpMRI: prepare for success. Clin Radiol. 2019;74(11):831–40. https://doi.org/10.1016/j.crad.2018.12.003.
Richenberg J, Logager V, Panebianco V, et al. The primacy of multiparametric MRI in men with suspected prostate cancer. Eur Radiol. 2019;29(12):6940–52. https://doi.org/10.1007/s00330-019-06166-z.
Zawaideh JP, Sala E, Shaida N, et al. Diagnostic accuracy of biparametric versus multiparametric prostate MRI: assessment of contrast benefit in clinical practice. Eur Radiol. 2020;30(7):4039–49. https://doi.org/10.1007/s00330-020-06782-0.
Tamada T, Kido A, Yamamoto A, et al. Comparison of biparametric and multiparametric MRI for clinically significant prostate cancer detection With PI-RADS Version 2.1. J Magn Reson Imaging 2021;53(1):283–91. https://doi.org/10.1002/jmri.27283.
Gibbs P, Liney GP, Pickles MD, et al. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest Radiol. 2009;44(9):572–6. https://doi.org/10.1097/RLI.0b013e3181b4c10e.
Sathiadoss P, Schieda N, Haroon M, et al. Utility of Quantitative T2-Map** Compared to Conventional and Advanced Diffusion Weighted Imaging Techniques for Multiparametric Prostate MRI in Men with Hip Prosthesis. J Magn Reson Imaging:10. doi: https://doi.org/10.1002/jmri.27803
Kitzing YX, Prando A, Varol C, et al. Benign conditions that mimic prostate carcinoma: MR imaging features with histopathologic correlation. Radiographics. 2016;36(1):162–75. https://doi.org/10.1148/rg.2016150030.
Rosenkrantz AB, Taneja SS. Radiologist, Be Aware: Ten Pitfalls That Confound the Interpretation of Multiparametric Prostate MRI. Am J Roentgenol. 2014;202(1):109–20. https://doi.org/10.2214/ajr.13.10699.
Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol. 2016;69(1):16–40. https://doi.org/10.1016/j.eururo.2015.08.052.
Le JD, Tan N, Shkolyar E, et al. Multifocality and prostate cancer detection by multiparametric magnetic resonance imaging: correlation with whole-mount histopathology. Eur Urol. 2015;67(3):569–76. https://doi.org/10.1016/j.eururo.2014.08.079.
Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol 2019;76(3):340–51. https://doi.org/10.1016/j.eururo.2019.02.033.
Sabouri S, Chang SD, Savdie R, et al. Luminal water imaging: a new MR Imaging T2 map** technique for prostate cancer diagnosis. Radiology. 2017;284(2):451–9. https://doi.org/10.1148/radiol.2017161687.
Chatterjee A, Bourne RM, Wang S, et al. Diagnosis of prostate cancer with noninvasive estimation of prostate tissue composition by using hybrid multidimensional MR imaging: a feasibility study. Radiology. 2018;287(3):864–73. https://doi.org/10.1148/radiol.2018171130.
Chatterjee A, Mercado C, Bourne RM, et al. Validation of prostate tissue composition by using hybrid multidimensional MRI: correlation with histologic findings. Radiology. 2022;302(2):368–77. https://doi.org/10.1148/radiol.2021204459.
Kim D, Doyle EK, Wisnowski JL, et al. Diffusion-relaxation correlation spectroscopic imaging: a multidimensional approach for probing microstructure. Magn Reson Med. 2017;78(6):2236–49. https://doi.org/10.1002/mrm.26629.
Zhang Z, Wu HH, Priester A, et al. Prostate microstructure in prostate cancer using 3-T MRI with diffusion-relaxation correlation spectrum imaging: validation with whole-mount digital histopathology. Radiology. 2020;296(2):348–55. https://doi.org/10.1148/radiol.2020192330.
Lee HJ, Hwang SI, Han S-M, et al. Image-based clinical decision support for transrectal ultrasound in the diagnosis of prostate cancer: comparison of multiple logistic regression, artificial neural network, and support vector machine. Eur Radiol. 2010;20(6):1476–84. https://doi.org/10.1007/s00330-009-1686-x.
Erickson BJ, Korfiatis P, Akkus Z, et al. Machine learning for medical imaging. Radiographics. 2017;37(2):505–15. https://doi.org/10.1148/rg.2017160130.
Wu M, Krishna S, Thornhill RE, et al. Transition zone prostate cancer: Logistic regression and machine-learning models of quantitative ADC, shape and texture features are highly accurate for diagnosis. J Magn Reson Imaging. 2019;50(3):940–50. https://doi.org/10.1002/jmri.26674.
Bernatz S, Ackermann J, Mandel P, et al. Comparison of machine learning algorithms to predict clinically significant prostate cancer of the peripheral zone with multiparametric MRI using clinical assessment categories and radiomic features. Eur Radiol. 2020;30(12):6757–69. https://doi.org/10.1007/s00330-020-07064-5.
Gholizadeh N, Simpson J, Ramadan S, et al. Voxel-based supervised machine learning of peripheral zone prostate cancer using noncontrast multiparametric MRI. J Appl Clin Med Phys. 2020;21(10):179–91. https://doi.org/10.1002/acm2.12992.
Li J, Weng Z, Xu H, et al. Support Vector Machines (SVM) classification of prostate cancer Gleason score in central gland using multiparametric magnetic resonance images: a cross-validated study. Eur J Radiol. 2018;98:61–7. https://doi.org/10.1016/j.ejrad.2017.11.001.
Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313(4):390–7. https://doi.org/10.1001/jama.2014.17942.
Calio BP, Sidana A, Sugano D, et al. Risk of upgrading from prostate biopsy to radical prostatectomy pathology-does saturation biopsy of index lesion during multiparametric magnetic resonance imaging-transrectal ultrasound fusion biopsy help? J Urol. 2018;199(4):976–82. https://doi.org/10.1016/j.juro.2017.10.048.
Pepe P, Pennisi M, Fraggetta F. How many cores should be obtained during saturation biopsy in the era of multiparametric magnetic resonance? experience in 875 patients submitted to repeat prostate biopsy. Urology. 2020;137:133–7. https://doi.org/10.1016/j.urology.2019.11.016.
Natarajan S, Marks LS, Margolis DJ, et al. Clinical application of a 3D ultrasound-guided prostate biopsy system. Urol Oncol. 2011;29(3):334–42. https://doi.org/10.1016/j.urolonc.2011.02.014.
Fleiss JL, Cohen J. The equivalence of weighted kappa and the intraclass correlation coefficient as measures of reliability. Educ Psychol Measur. 1973;33(3):613–9. https://doi.org/10.1177/001316447303300309.
Sadinski M, Karczmar G, Peng Y, et al. Pilot study of the use of hybrid multidimensional T2-weighted imaging-DWI for the diagnosis of prostate cancer and evaluation of gleason score. AJR Am J Roentgenol. 2016;207(3):592–8. https://doi.org/10.2214/AJR.15.15626.
Wang S, Peng Y, Medved M, et al. Hybrid multidimensional T(2) and diffusion-weighted MRI for prostate cancer detection. J Magn Reson Imaging. 2014;39(4):781–8. https://doi.org/10.1002/jmri.24212.
Chatterjee A, Harmath C, Oto A. New prostate MRI techniques and sequences. Abdom Radiol (NY). 2020;45(12):4052–62. https://doi.org/10.1007/s00261-020-02504-8.
Syversen IF, Elschot M, Sandsmark E, et al. Exploring the diagnostic potential of adding T2 dependence in diffusion-weighted MR imaging of the prostate. PLoS ONE. 2021;16(5): e0252387. https://doi.org/10.1371/journal.pone.0252387.
Henry GH, Malewska A, Joseph DB, et al. A cellular anatomy of the normal adult human prostate and prostatic urethra. Cell Rep. 2018;25(12):3530-42 e5. https://doi.org/10.1016/j.celrep.2018.11.086.
Mai Z, Zhou Z, Yan W, et al. The transverse and vertical distribution of prostate cancer in biopsy and radical prostatectomy specimens. BMC Cancer. 2018;18(1):1205. https://doi.org/10.1186/s12885-018-5124-9.
Acknowledgements
I wish to thank Guiqin Liu for advice and encouragement on the study.
Funding
Science and Technology Commission of Shanghai Municipality; Contract grant number: 18DZ1930104; Shanghai Jiao Tong University medical-engineering cross fund; Contract grant number: YG2021QN27.
Author information
Authors and Affiliations
Contributions
**aobin Wei, Li Zhu and Yanyan Zeng contributed equally to this work. **aobin Wei contributed to conceptualization, data curation, formal analysis, writing-original draft, and writing-review & editing. Li Zhu contributed to data curation and formal analysis. Yanyan Zeng, Ke Xue and Yongming Dai contributed to Investigation, Validation, Visualization, Software, Visualization. Jianrong Xu and Guiqin Liu contributed to Writing-original draft and Writing-review. Fang Liu contributed to revising the manuscript. Wei Xue, Dongmei Wu and Guangyu Wu cocorresponding to this work. We certify that we have participated sufficiently in the work to take public responsibility for the appropriateness of the study design and method and the collection, analysis, and interpretation of the data. All authors have reviewed the final version of the manuscript and approve it for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of our institute (Approval No. KY2018-213). Written informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wei, X., Zhu, L., Zeng, Y. et al. Detection of prostate cancer using diffusion-relaxation correlation spectrum imaging with support vector machine model – a feasibility study. Cancer Imaging 22, 77 (2022). https://doi.org/10.1186/s40644-022-00516-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40644-022-00516-9