Abstract
This survey explores how Deep Learning has battled the COVID-19 pandemic and provides directions for future research on COVID-19. We cover Deep Learning applications in Natural Language Processing, Computer Vision, Life Sciences, and Epidemiology. We describe how each of these applications vary with the availability of big data and how learning tasks are constructed. We begin by evaluating the current state of Deep Learning and conclude with key limitations of Deep Learning for COVID-19 applications. These limitations include Interpretability, Generalization Metrics, Learning from Limited Labeled Data, and Data Privacy. Natural Language Processing applications include mining COVID-19 research for Information Retrieval and Question Answering, as well as Misinformation Detection, and Public Sentiment Analysis. Computer Vision applications cover Medical Image Analysis, Ambient Intelligence, and Vision-based Robotics. Within Life Sciences, our survey looks at how Deep Learning can be applied to Precision Diagnostics, Protein Structure Prediction, and Drug Repurposing. Deep Learning has additionally been utilized in Spread Forecasting for Epidemiology. Our literature review has found many examples of Deep Learning systems to fight COVID-19. We hope that this survey will help accelerate the use of Deep Learning for COVID-19 research.
Similar content being viewed by others
Introduction
SARS-CoV-2 and the resulting COVID-19 disease is one of the biggest challenges of the 21st century. At the time of this publication, about 43 million people have tested positive and 1.2 million people have died as a result [1]. Fighting this virus requires heroism of healthcare workers, social organization and technological solutions. This survey focuses on advancing technological solutions, with an emphasis on Deep Learning. We additionally highlight many cases where Deep Learning can facilitate social organization such as Spread Forecasting, Misinformation Detection, or Public Sentiment Analysis. Deep Learning has gained massive attention by defeating the world champion at Go [2], controlling a robotic hand to solve a Rubik’s cube [3], and completing fill-in-the-blank text prompts [4]. Deep Learning is advancing very quickly, but what is the current state of this technology? What problems does Deep Learning have the capability of solving? How do we articulate COVID-19 problems for the application of Deep Learning? We explore these questions through the lens of Deep Learning applications fighting COVID-19 in many ways.
This survey aims to illustrate the use of Deep Learning in COVID-19 research. Our contributions are also follows:
-
This is the first survey viewing COVID-19 applications solely through the lens of Deep Learning. In comparison with other surveys on COVID-19 applications in Data Science or Machine Learning, we provide extensive background on Deep Learning.
-
For each application area surveyed, we provide a detailed analysis of how the given data is inputted to a deep neural network and how learning tasks are constructed.
-
We provide an exhaustive list of applications in data domains such as Natural Language Processing, Computer Vision, Life Sciences, and Epidemiology. We particularly focus on work in Literature Mining for COVID-19 research papers, compiling papers from the ACL 2020 NLP-COVID workshop.
-
Finally, we review common limitations of Deep Learning including Interpretability, Generalization Metrics, Learning from Limited Labeled Data, and Data Privacy. We describe how these limitations impact each of the surveyed COVID-19 applications. We additionally highlight research tackling these issues.
Our survey is organized into four primary sections. We start with a “Background” on Deep Learning to explain the relationship with other Artificial Intelligence technologies such as Machine Learning or Expert Systems. This background also provides a quick overview of SARS-CoV-2 and COVID-19. The next section lists and explains “Deep Learning applications for COVID-19”. We organize surveyed applications by input data type, such as text or images. This is different from other surveys on COVID-19 that organize applications by scales such as molecular, clinical, and society-level [5, 6].
From a Deep Learning perspective, organizing applications by input data type will help readers understand common frameworks for research. Firstly, this avoids repeatedly describing how language or images are inputted to a Deep Neural Network. Secondly, applications working with the same type of input data have many similarities. For example, cutting-edge approaches to Biomedical Literature Mining and Misinformation Detection both work with text data. They have many commonalities such as the use of Transformer neural network models and reliance on a self-supervised representation learning scheme known as language modeling. We thus divide surveyed COVID-19 applications into “Natural Language Processing”, “Computer Vision”, “Life Sciences”, and “Epidemiology”. However, our coverage of applications in Life Sciences diverges from this structure. In the scope of Life Sciences, we describe a range of input data types, such as tabular Electronic Health Records (EHR), textual clinical notes, microscopic images, categorical amino acid sequences, and graph-structured network medicine.
The datasets used across these applications tend to share the common limitation of size. In a rapid pandemic response situation, it is especially challenging to construct large datasets for Medical Image Analysis or Spread Forecasting. This problem is evident in Literature Mining applications such as Question Answering or Misinformation Detection as well. Literature Mining data is an interesting situation for Deep Learning because we have an enormous volume of published papers. Despite having such a large unlabeled dataset, downstream applications such as question answering or fact verification datasets are extremely small in comparison. We will continually discuss the importance of pre-training for Deep Learning. This paradigm relies on either supervised or self-supervised transfer learning. Of core importance, explored throughout this paper, is the presence of in-domain data. Even if it is unlabeled, such as a biomedical literature corpus, or slightly out-of-domain, such as the CheXpert radiograph dataset for Medical Image Analysis [7], availability of this kind of data is paramount for achieving high performance.
When detailing each application of Deep Learning to COVID-19, we place an emphasis on the representation of data and the task. Task descriptions mostly describe how a COVID-19 application is constructed as a learning problem. We are solely focused on Deep Learning applications, and thus we are referring to representation learning of raw, or high-dimensional data. A definition and overview of representation learning is provided in “Background” section. The following list quickly describes different learning variants found in our surveyed applications:
-
Supervised Learning optimizes a loss function with respect to predicted and ground truth labels. These ground truth labels require manual annotation.
-
Unsupervised Learning does not use labels. This includes clustering algorithms that look for intrinsic structure in data.
-
Self-Supervised Learning optimizes a loss function with respect to the predicted and ground truth labels. Differently from Supervised Learning, these labels are constructed from a separate computing process, rather than human annotation.
-
Semi-Supervised Learning uses a mix of human labeled and unlabeled data for representation learning.
-
Transfer Learning describes initializing training with the representation learned from a previous task. This previous task is most commonly ImageNet-based supervised learning in “Natural Language Processing” or Internet-scale language modeling in “Computer Vision”.
-
Multi-Task Learning simultaneously optimizes multiple loss function, usually either interleaving updates or applying regularization penalties to avoid conflicting gradients from each loss.
-
Weakly Supervised Learning refers to supervised learning with heuristically labeled data, rather than carefully labeled data.
-
Multi-Modal Learning describes representation learning in multiple data types simultaneously, such as images and text or images and electronic health records.
-
Reinforcement Learning optimizes a loss function with respect to a series of state to action predictions. This is especially challenging due to credit assignment in the sequence of state to action map**s when receiving sparse rewards.
It is important to note the distinction between these learning task constructions in each of our surveyed applications. We further contextualize our surveyed applications with an overview of “Limitations of Deep Learning”. These limitations are non-trivial and present significant barriers for Deep Learning to fight COVID-19 related problems. Solutions to these issues of “Interpretability”, “Generalization metrics”, “Learning from limited labeled datasets”, and “Data privacy” will be important to many applications of Deep Learning. We hope describing how the surveyed COVID-19 applications are limited by these issues will develop intuition about the problems and motivate solutions. Finally, we conclude with a “Discussion” and “Conclusion” from our literature review. Our Discussion describes lessons learned from a comprehensive literature review and plans for future research.
Deep Learning for “Natural Language Processing” (NLP) has been extremely successful. Applications for COVID-19 include Literature Mining, Misinformation Detection, and Public Sentiment Analysis. Searching through the biomedical literature has been extremely important for drug repurposing. A success case of this is the repurposing of baricitinib [8], an anti-inflammatory drug used in rheumatoid arthritis. The potential efficacy of this drug was discovered by querying biomedical knowledge graphs. Modern knowledge graphs utilize Deep Learning for automated construction. Other biomedical literature search systems use Deep Learning for information retrieval from natural language queries. These Literature Mining systems have been extended with question answering and summarization models that may revolutionize search altogether. We additionally explore how NLP can fight the “infodemic” by detecting false claims and presenting evidence. NLP is also useful to evaluate public sentiment about the pandemic from data such as tweets and provide tools for social scientists to analyze free-text response surveys.
“Computer Vision” is another mature application domain of Deep Learning. The Transformer revolution in Natural Language Processing largely owes its success to Computer Vision’s pioneering into large datasets, massive models, and the utilization of hardware that accelerates parallel computation, namely GPUs [Epidemiology”. How many people do we expect to be infected with COVID-19? How long do we have to quarantine for? These are query examples for our search systems, described as NLP applications, but epidemiological models are the source of these answers. We will begin exploring this through the lens of “black-box” forecasting models that look at the history of infections and other information to predict into the future. We will then look at SIR models, a set of differential equations modeling the transition from Susceptible to Infected to Recovered. These models find the reproductive rate of the virus, which can characterize the danger of letting herd immunity develop naturally. To formulate this as a Deep Learning task, a Deep Neural Network approximates the time-varying strength of quarantine in the SIR model, since integrating the Exposed population would require extremely detailed data. Parameter optimizers from Deep Learning such as Adam [11] can be used to solve differential equations as well. We briefly investigate the potential of Contact Tracing data. Tracking the movement of individuals when they leave their quarantine could produce incredibly detailed datasets about how the virus spreads. We will explore what this data might look like and some tasks for Deep Learning. This is another application area that relates heavily to our discussion of data privacy in “Limitations of Deep Learning”.
These applications of Deep Learning to fight COVID-19 are promising, but it is important to be cognizant of the drawbacks to Deep Learning. We focus on the issues of “Interpretability”, “Generalization metrics”, “Learning from limited labeled datasets”, and “Data Privacy”. It is very hard to interpret the output of current Deep Learning models. This problem is further compounded by the lack of a reliable measure of uncertainty. When the model starts to see out-of-distribution examples, data points sampled from a different distribution than the data used to train the model, most models will continue to confidently misclassify these examples. It is very hard to categorize how well a trained model will generalize to new data distributions. Furthermore, these models can fail at simple commonsense tasks, even after achieving high performance on the training dataset. Achieving this high performance in the first place comes at the cost of massive, labeled datasets. This is unpractical for most clinical applications like Medical Image Analysis, as well as for quickly building question-answering datasets. Finally, we have to consider data privacy with these applications. Will patients feel comfortable allowing their ICU activity to be monitored by an intelligent camera? Would patients be comfortable with their biological data and medical images being stored in a central database for training Deep Learning models? This introduction should moderate enthusiasm about Deep Learning as a panacea to all problems. However, we take an optimistic look at these problems in our section “Limitations of Deep Learning”, explaining solutions to these problems as well, such as self-explanatory models or federated learning.
Background
This section will provide a background for this survey. We begin with a quick introduction to COVID-19, followed by what Deep Learning is and how it relates to other Artificial Intelligence technologies. Finally, we present the relationship of this survey with other works reviewing the use of Artificial Intelligence, Data Science, or Machine Learning to fight COVID-19.
SARS-CoV-2 originated from Wuhan, China and spread across the world, causing a global pandemic. The response has been a mixed bag of mostly chaos and a little optimism. Scientists were quick to sequence and publish the complete genome of the virus [12], and individuals across the world quarantined themselves to contain the spread. Scientists have lowered barriers for collaboration. However, there have been many negative issues surrounding the pandemic. The quick infection and lack of resources has overloaded hospitals and heavily burdened healthcare workers. SARS-CoV-2 has a unique characteristic of peak infection before symptom manifestation that has worked in the favor of the virus. Misinformation has spread so rampantly, a new field of “infodemiology” has sprouted to fight the “infodemic”. Confusion of correct information is compounded by a rapidly growing body of literature surrounding SARS-CoV-2 and COVID-19. This research is emerging very quickly and new tools are needed to help scientists organize this information.
A technical definition of Deep Learning is the use of neural networks with more than one or two layers. A neural network “layer” is typically composed of a parametric, non-linear transformation of an input. Stacking these transformations forms a statistical data structure capable of map** high-dimensional inputs to outputs. This map** is executed by optimizing the parameters. Gradient descent is the tool of choice for this optimization. Gradient descent works by taking the partial derivative of the loss function with respect to each of the parameters, and updating the parameters to minimize the loss function. Deep Learning gets the name “Deep” in reference to stacking several of these layers. The second part of the name, “Learning”, references parameter optimization. State-of-the-art Deep Learning models typically contain between 100 million to 10 billion parameters and 50–200 layers. Two of the largest models publicly reported are composed of 600 and 175 billion parameters [43]. The TREC-COVID dataset consists of topics where each topic is composed of a query, question, and narrative. The narrative is a longer description of the question. Figure 3 shows an example of the interface the authors use to label the TREC-COVID dataset.

(Image taken from Voorhees et al. [43])
Interface for human labeling TREC-COVID documents
The following list provides a quick description of some Literature Mining systems built from datasets such as CORD-19 and TREC-COVID. These systems use a combination of Information Retrieval, Knowledge Graph Construction, Question Answering, and Summarization to facilitate exploration into the COVID-19 scientific literature.
-
CO-Search [38] is a Retrieve-then-Rank system composed of many parts, shown in Fig. 4. Before answering any user queries, the entire document corpus is encoded with Sentence-BERT (SBERT) [44], TF-IDF, and BM25 features. A user enters a query and it is encoded with a similar combination of featurizers. This query encoding is used to index the featurized documents and thus return the most semantically similar documents to the query. Having retrieved these documents, the next task is ranking for presentation to the user. First, the retrieved documents and query are passed as input to a multi-hop question answering model and an abstractive summarization system. The output from these models are weighted with the original scoring from the retrieval step, and the top scoring documents are presented to answer the query.
CO-Search is a combination of many cutting-edge NLP models. Their pre-training task for the SBERT encoder is very interesting. The authors train SBERT to take a paragraph from a research paper and classify whether it cites another paper, given only the title of the other paper. SBERT is a siamese architecture which takes one sequence as input at a time. SBERT uses the cosine similarity loss between the output representation of each separately encoded sequence to compare the paragraph and the title. The representation learned from this pre-training task is then used to encode the documents and queries, as previously describe. We will unpack the question answering and abstractive summarization systems later in the survey.
Fig. 4 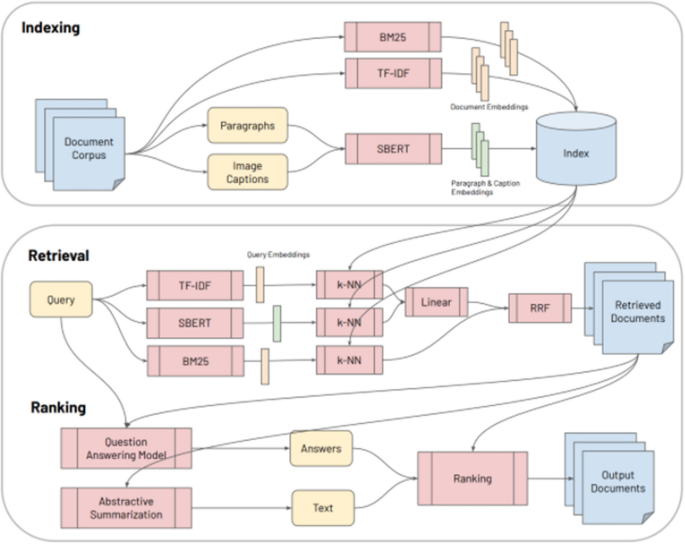
(Image taken from Esteva et al. [38])
CO-Search System Architecture
-
Covidex [45] is a Retrieve-then-Rank system combining keyword retrieval with neural re-ranking. The most different aspects of Covidex as compared to CO-Search are a sequence-to-sequence (seq2seq) approach to re-ranking and bootstraps the training of this model from the MS MARCO [46] passage ranking dataset. The MS MARCO dataset contains 8.8M passages obtained from the top 10 results by the Bing search engine, corresponding to 1M unique queries. The monoT5 [47] seq2seq re-ranker takes as input “Query q: Document: d Relevant: “ to classify whether the document is relevant or not to the query.
-
SLEDGE [48] deploys a similar pipeline of keyword-based retrieval followed by neural ranking. Differently from Covidex, SLEDGE utilizes ths SciBERT [27] model for re-ranking. SciBERT is an extension to BERT that has been pre-trained on Scientific Text. Additionally, the authors of SLEDGE find large gains by integrating the publication date of the articles into the input representation.
-
CAiRE-COVID [39] is a similar system to CO-Search. The primary difference comes from the use of the MRQA [49] model and avoiding fine-tuning the QA models on COVID-19 related datasets. This system tests the generalization of existing QA models comprising of a pre-trained BioBERT [26] fine-tuned on the SQuAD dataset. The user interface of CAiRE-COVID is depicted in Fig. 5
Fig. 5 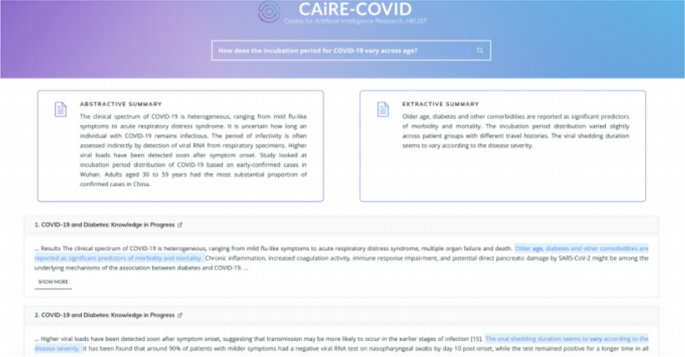
(Image taken from Su et al. [39])
User Interface for the CAiRE-COVID Literature Mining system consisting of Extractive Summary, Abstractive Summary, and most relevant documents
The preceding list are examples of Information Retrieval (IR) systems. As mentioned previously, IR describes the task of finding relevant documents from a large set of candidates given a query. These systems typically deploy multi-stage processing to break up the computational complexity of the problem. The first stage is the retrieval stage, where a set of documents much smaller than the total set is returned that best match the query. This first stage of retrieval has only recently integrated the use of neural representations. As previously listed, many systems combine TF-IDF or BM25 sparse vectors with dense representations derived from SBERT. The relationship between these representations is well stated in Karpukhin et al. [51] describe the challenge of constructing datasets for COVID-19 QA in a similar format as the SQuAD dataset. Five annotators working on constructing this dataset for 23 hours resulted in 124 question-article pairs. This includes deconstruction of topics such as “decontamination based on physical science” into multiple questions such as “UVGI intensity used for inactivating COVID-19” and “Purity of ethanol to inactive COVID-19”. The authors demonstrate the zero-shot capabilities of pre-trained language models on these questions. This is an encouraging direction as Shick and Shutze [52, 53] have recently shown how to perform few-shot learning with smaller language models.
Knowledge Graph Construction
One of the best mechanisms of organizing information is the use of Knowledge Graphs. Figure 6 is an example of a Knowledge Graph of our surveyed Deep Learning applications for COVID-19. Each relation in this example is A “contains” B. This is an illustrative example of organizing information topologically. It is often easier to understand how complex systems work or ideas connect when explicitly linked and visualized this way. We want to construct these kinds of graphs with all of the biomedical literature relevant to COVID-19. This raises questions about the construction and usability of knowledge graphs. Can we automatically construct these graphs from research papers and articles? At what scale is this organization too overwhelming to look through?
In application to COVID-19, we would like to construct Biomedical Knowledge Graphs. These graphs capture relations between entities such as proteins and drugs and how they are related such as “chemical A inhibits the binding of protein B”. Richardson et al. [8] describe how they use the BenevolentAI knowledge graph to discover baricitinib as potential treatment for COVID-19. In this section, we focus on how NLP is used to construct these graphs. Under our “Life Sciences” section, we will discuss the potential use of graph neural networks to mine information from the resulting graph-structured data.
Figure 7 shows examples of different nodes and links. We see the 2019-nCoV node (continually referred to as COVID-19 in our survey), the ACE2 membrane protein node, and the Endocytosis cellular process node, to name a few. The links describe how these different nodes are related such as 2019-nCoV “Binds” ACE2, ACE2 “Expressed in” AT2 lung cell. Richardson et al. [8] describe how this structure allows them to query the graph and form hypotheses about approved drugs. The authors originally have the hypothesis that the AAK1 protein is one of the most known regulators of endocytosis and that disruption of this might stop the virus from entering the cells. The knowledge graph returns 378 AAK1 inhibitors, 47 having been approved for medical use and 6 that have inhibited AAK1 with high affinity. However, the knowledge graph shows that many of these compounds have serious side-effects. baricitinib, one of the 6 AAK1 inhibitors, also binds another kinase that regulates endocytosis. The authors reason that this can reduce both viral entry and inflammation in patients. This is further described in Stebbing et al. [54].

(Image taken from Richardson et al. [8])
BenevolentAI Knowledge Graph used to suggest baricitinib as a treatment for COVID-19
We note that this kind of traversal of the Knowledge Graph requires significant prior knowledge on the part of the human-in-the-loop. This motivates our emphasis on Human-AI interaction in “Limitations of Deep Learning”. Without human accessible interfaces, the current generation of Artificial Intelligence systems are useless. We will describe how Deep Learning, rather than expert querying, can be used to mine these graphs in our section on “Life Sciences”. Within the scope of NLP, we describe how these graphs are constructed from large datasets. This is referred to as Automated Knowledge Base Construction.
A Knowledge Graph is composed of a set of nodes and edges. We can set this up as a classification task where Deep Learning models are tasked to classify nodes in a body of text. This task is known as Named Entity Recognition (NER). In addition to identifying nodes, we want to classify their relation. As a supervised learning task, these labels include the set of edges we want our Knowledge Graph to contain, such as “inhibits” or “binds to”. Deep Learning serves classify nodes and edges according to a human-designed set, not to define new nodes and relations. Describing their system called PaperRobot, built prior to the pandemic outbreak, Wang et al. describe that “creating new nodes usually means discovering new entities (e.g. new proteins) through a series of laboratory experiments, which is probably too difficult for PaperRobot” [55].
The labeled nodes are linked together with an entity ontology defined by biological experts. Defined by Guarino et al., “computational ontologies are a means to formally model the structure of a system, i.e., the relevant entities and relations that emerge from its observation, and which are useful to our purposes” [56]. Wang et al. [57] link the MeSH IDs entities together based on the Comparative Toxicogenomics Database (CTD) ontology. From Davis et al. [58], “CTD is curated by professional biocurators who leverage controlled vocabularies, ontologies, and structured notation to code a triad of core interactions describing chemical-gene, chemical-disease and gene-disease relationships”.
Wise et al. [59] construct a similar graph containing 336,887 entities and 3,332,151 relations. The set of nodes and edges are shown in Fig. 8. The authors use a combination of graph and semantic embeddings to answer questions with the top-k most similar articles, similar to the Information Retrieval systems previously described. Hosted on http://www.CORD19.aws, their system has seen over 15 million queries across more than 70 countries.

(Image taken from Wise et al. [60])
Meta-data on the count of Entities in CKG and Relation information
Zeng et al. [61] describe the construction of a more ambitious Knowledge Graph from a large scientific corpus of 24 million PubMed publications. Their graph contains 15 million edges across 39 types of relationships connecting drugs, diseases, proteins, genes, pathways, and expression. From their graph they propose 41 repurposable drugs for COVID-19. Chen et al. [62] present a more skeptical view of NER for automated COVID-19 knowledge graph construction. They highlight that even the more in-domain BioBERT model has not been trained on enough data to recognize entities such as “Thrombocytopenia”, or even “SARSCOV-2”. They instead use a co-occurrence frequency to extract nodes and use word2vec [63] similarity to filter edges.
In SciSight [64], the authors design a knowledge graph that is integrated with the social dynamics of scientific research. They motivate their approach highlighting that most of these Literature Mining systems are designed for targeted search. Targeted search is defined as search where the researchers know what they are looking for. SciSight is designed for exploration and discovery. This is done by the construction of knowledge graph of topics as well as the social graphs of the researchers themselves.
Misinformation Detection
The spread of information related to SARS-CoV-2 and COVID-19 has been chaotic, denoted as an infodemic [65]. From conspiracy theories ranging from causal attribution of 5G networks to false treatments and reporting of scientific information, how can Deep Learning be used to fight the infodemic? In our survey we will look at this under the lens of the spread of misinformation and the detection of it. The detection of misinformation has been formulated as a text classification or semantic similarity problem. Our original description of the GLUE benchmark should help readers understand the core Deep Learning problem in the following surveyed experiments.
Many studies have built classification models to flag tweets potentially containing misinformation. These papers mostly differ in how they label these tweets. Alam et al. [66] label tweets according to 7 question labels; contains a verifiable factual claim, is likely to contain false information, is of interest to the general public, is potentially harmful to a person, a company, a product, or society, requires verification by a fact-checker, poses a specific kind of harm to society, and requires the attention of a government entity. Dharawat et al. [67] look at the seriousness of misinformation, reasoning that “urging users to eat garlic is less severe than urging users to drink bleach”. Their Covid-HeRA dataset contains 61,286 tweets labeled as not severe, possibly severe, highly severe, refutes/rebuts, and real news/claims. Hossain et al. [68] collaborate with researchers from the UCI school of medicine to establish a set of common Misconceptions. These misconceptions are used to label Tweets. Examples of this are shown in Fig. 9.

(Image taken from Hossain et al. [69])
Examples of Misinformation Labels
The detection of misinformation and fact verification has been studied before the COVID-19 infodemic. The most notable dataset of this is the FEVER, Fact Extraction and Verification, dataset [70]. This dataset contains 185,445 claims generated by human annotators. The annotators of the dataset were presented an introduction section of a Wikipedia article and asked to generate a factual claim and then perturb that claim such that it is no longer factually verified. We refer readers to their paper to learn about additional challenges of constructing this kind of dataset [70].
Constructing a new dataset that properly frames a new task is a common theme in Deep Learning. Wadden et al. [71] construct the SciFact dataset to extend the ideas of FEVER to COVID-19 applications. This is different from classification task formulations previously mentioned in that they are more creative in task design. SciFact and FEVER introduce new datasets that show how supervised learning can tackle Misinformation Detection. Wadden et al. [71] design the SciFact dataset to not only classify a claim as true or false, but to provide supporting and refuting evidence as well. Wadden et al. [71] deploy a clever annotation scheme of using “citances”, sentences in scientific papers that cite another paper, as examples of supporting or refuting evidence for a claim. Examples of this are shown in Fig. 10. The baseline system they deploy to test the dataset is an information retrieval model. We refer readers to our previous section on Literature Mining to learn more about these models.

(Image taken from Wadden et al. [71])
COVID-19 claim examples about COVID-19 and corresponding evidence retrieved
The authors of SciFACT and FEVER baseline their datasets with neural information retrieval systems. Their systems rely on sparse feature vectors such as TF-IDF and distributed representations of knowledge implicitly stored in neural networks weights. In the previous section we described Knowledge Graphs. Knowledge Graphs for fact verification may be more reliable than neural network systems. We could additionally keep an index of sentences and passages that resulted in relations added to the knowledge graph. This could facilitate evidence explanations. The primary difference with this approach is the extent of automation. Querying Knowledge Base and sifting through relational evidence requires much more human-in-the-loop interaction than neural systems. This approach is also bottlenecked by the problem of scale with evidence. We will continue to compare the prospects of Knowledge Graphs and Neural Systems in our “Discussion” section.
Public Sentiment Analysis
The uncertainty of COVID-19 and the challenge of quarantine ignited mental health issues for many people. NLP can help us gauge how the public is faring from multiple angles such as economic, psychological, and sociological analysis. Are individuals endorsing or rejecting health behaviors which help reduce the spread of the virus? Previous studies have looked at the use of Twitter data for election sentiment [72, 73]. This section covers extensions of this work looking into aspects of COVID-19.
Twitter is one of the primary sources of data for Public Sentiment Analysis. Other public data sources include news articles or Reddit. Tweet classification is very similar to the Single-Sentence GLUE benchmark [29] tasks previously described. Each Tweet has a maximum of 280 characters, which will easily fit into Transformer Neural Networks. Compared with Literature Mining, there is no need to carefully construct atomic units for Tweets.
The core question with Twitter data analysis is filtering of the extracted Tweets into categories. This is usually done by keyword matching. Muller et al. [28] use the keywords “wuhan”, “ncov”, “coronavirus”’, “covid”, “sars-cov-2”. Filtering by these keywords over a span from January 12th to April 16th, 2020 resulted in 22.5 M collected tweets. The authors use this dataset to train COVID-Twitter-BERT with self-supervised language modeling. Previously, we discussed the benefit of in-domain data for self-supervised pre-training detailed in [23]. COVID-Twitter-BERT is then fine-tuned for five different Sentiment classification datasets. Two of these datasets target Vaccine Sentiment and Maternal Vaccine Stance. Compared to fine-tuning the BERT [24] model pre-trained on out-of-domain data sources such as Wikipedia and books, fine-tuned COVID-Twitter-BERT models achieve an average 3% improvement across 5 classification tasks. The average improvement was brought down by a very small improvement on the Stanford Sentiment Treebank 2 (SST-2) dataset, which does not consist of Tweets. This further highlights the benefits of in-domain self-supervised pre-training for Natural Language Processing, and more broadly, Deep Learning applications.
Nguyen et al. [74] construct a dataset of 10K COVID tweets. These tweets are labeled as to whether they provide information about recovered, suspected, confirmed, and death cases, as well as location or travel history of the cases, or if they are uninformative altogether. This dataset was used in the competition WNUT-2020 Task 2: Identification of Informative COVID-19 English Tweets. Chauhan [75] describes the efficacy of data augmentation to prevent over-reliance on easy clues such as “deaths” and “died” to identify informative tweets. Sancheti et al. [76] describe the use of semi-supervised learning for this task, finding benefits from leveraging unlabeled tweets.
Loon et al. [77] explore the notion that SARS-CoV-2 “has taken on heterogeneous socially constructed meanings, which vary over the population and shape communities’ response to the pandemic.” They tie Twitter data with Google COVID-19 Community Mobile Reports [78] and find that political sentiment can predict how much people are social distancing. They find that residents social distanced less in areas where the COVID sentiment endorsed concepts of fraud, the political left, and more benign illnesses.
Further exploration into Deep Learning components such as architecture design, loss functions, or activations [79] will not be as important as dataset curation. With a strong dataset, text classification is an easy task for cutting-edge Transformer models. Later on, we will look at the Limitations of Deep Learning that highlight what makes this task challenging from a perfect performance perspective. Another topic in our coverage of limitations is the importance of Human-AI interaction. This is relevant for all applications discussed, but especially for public sentiment with respect to user interfaces. Public sentiment is typically interpreted by economists, psychologists, and sociologists who may not be comfortable adding desired functionality to a PyTorch [80] or Tensorflow [81] codebase. We emphasize the importance of user interfaces that allow users to integrate Public Sentiment Analysis into free-text answers in surveys.
Computer Vision
Computer Vision, powered by Deep Learning, has a very impressive resume. In 2012, AlexNet implemented a Convolutional Neural Network with GPU optimizations. AlexNet set its sights on the ImageNet dataset. The result significantly past the competition with a 63.3% top-1 accuracy. This marked a large improvement from 50.9% with manually engineered image features. AlexNet inspired further interest in Deep Neural Networks for Computer Vision. Researchers designed new architectures, new ways of representation learning from unlabeled data, and new infrastructure for training larger models. In 2020, eight years after AlexNet, the Noisy Student EfficientNet-L2 model reached 88.4% top-1 accuracy, an absolute improvement of 25.1%. The Deep Computer Vision resume continues with generation of photorealistic facial images, transferring artistic style from one image to another, and enabling robotic control solely from visual input.
The success of Deep Computer Vision is largely attributed to the ImageNet dataset [82]. The ImageNet competition is a dataset that contains 1.2 million images labeled in 1,000 categories. Images are inputted to Deep Neural Networks as tensors of the dimension height × width × channels. For example, most ImageNet images have the dimension 128 × 128 × 3 pixels. The resolution of image inputs to Deep Learning is an important consideration for the sake of computational and storage cost.
Computer Vision stands to transform Healthcare in many ways. The most salient and frequently discussed application to COVID-19 is medical image diagnosis. Deep Learning has performed extremely well at medical image diagnosis and has underwent clinical trials across many diseases [83]. Medical image tasks mostly consider classification and segmentation, as well as reconstruction from 2-D slices in a CT-scan to make up the final 3-D view.
Computer Vision also stands to aid in subtle hospital operations. Haque et al. [10] describe “Ambient Intelligence” where Computer Vision aids in physical therapy to combat ICU-acquired weakness, ensures workers and patients wash their hands properly, and improves surgical training, to name a few. Computer Vision equips cameras to “understand” what they are recording. They can identify people. They can label every pixel of a road for the sake of self-driving cars. They can map satellite images into road maps. They can imagine what a sketch drawing would look like if it was a photographed object. Here we use “understand” for the sake of hyperbole, really meaning that it can answer semantic questions about image or video data. These applications describe the potential of Computer Vision to enable a large set of subtle applications in pandemic response, such as face mask detection and monitoring social distancing or hospital equipment inventory. We hope to excite readers about the implications and gravity of this technology; however, in “Limitations of Deep Learning” we highlight “Data Privacy” issues with this kind of surveillance.
Economic damage is one of the greatest casualties of COVID-19. How can we engineer contact-free manufacturing and supply chain processes that don’t endanger workers? Vision-based robotics is a promising direction for this. Current industrial robotics rely on solving differential equations to explicitly control desired movement. This works great when the input can be extremely controlled, but breaks down when the robot must generalize to new inputs. Vision-based robotics offers a more general control solution that can adapt to novel orientations of objects or weight distributions of boxes.
Medical Image Analysis
Assisting and automating Medical Image Analysis is one of the most commonly discussed applications of Deep Learning. These systems are improving rapidly and have moved into clinical trials. We refer readers to Topol’s guidelines for AI clinical research, exploring lessons from many clinical fields [84]. Our coverage of Medical Image Diagnosis for COVID-19 is mostly focused on classification of COVID-19 pneumonia, viral pneumonia, or healthy chest radiographs. There are many studies that focus on the Semantic Segmentation task, where every pixel in an image is classified. We discuss the Semantic Segmentation application in our section on “Interpretability” with respect to improving Human-AI Interaction. In our analysis, chest radiographs are sourced from either X-ray imaging or higher-resolution CT scans. Motivating the use of radiograph diagnosis, Fang et al. [1] find a 98% sensitivity with CT detection compared to 71% with RT-PCR. Ai et al. [109]. This describes whether a robot would be able to walk into any room and make a cup of coffee. We note that this level of generalization is unnecessary for most applications for COVID-19. We discuss this further in Generalization Metrics.
The range of robotic applications considered varies enormously in the complexity of manipulation required. For example, a robot unleashing a disinfecting spray on a room does not need to have fine-grained manipulation skills. However, a robot assisting a patient with a respiratory does. In a similar way, robots facilitating manufacturing and economic activity vary along this scale as well. Sorting bottles requires more vision than dexterity, whereas assembly requires incredibly dexterous manipulation.
Barfoot et al. [151].
Learning from limited labeled datasets
Availability of data and materials
Not applicable.
References
Worldometers. https://www.worldometers.info/coronavirus/. Accessed Jan 2021.
Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, Schrittwieser J, Antonoglou I, Panneershelvam V, Lanctot M, Dieleman S, Grewe D, Nham J, Kalchbrenner N, Sutskever I, Lillicrap T, Leach M, Kavukcuoglu K, Graepel T, Hassabis D. Mastering the game of go with deep neural networks and tree search. Nature. 2016;529:484–503.
OpenAI Akkaya I, Andrychowicz M, Chociej M, Litwin M, McGrew B, Petron A, Paino A, Plappert M, Powell G, Ribas R, Schneider J, Tezak N, Tworek J, Welinder P, Weng L, Yuan Q, Zaremba W, Zhang L. Solving Rubik’s cube with a robot hand; 2019. ar** the landscape of artificial intelligence applications against COVID-19; 2020. ar**v:2003.11336.
Latif S, Usman M, Manzoor S, Iqbal W, Qadir J, Tyson G, Castro I, Razi A, Kamel Boulos M, Crowcroft J. Preprint: Leveraging data science to combat COVID-19: a comprehensive review; 2020. https://doi.org/10.13140/RG.2.2.12685.28644/4.
Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, Seekins J, Mong DA, Halabi SS, Sandberg JK, Jones R, Larson DB, Langlotz CP, Patel BN, Lungren MP, Ng AY. CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison; 2019. ar**v:1901.07031.
Richardson P, Griffin I, Tucker C, Smith D, Oechsle O, Phelan A, Stebbing J. Baricitinib as potential treatment for 2019-ncov acute respiratory disease. Lancet. 2020. https://doi.org/10.1016/S0140-6736(20)30304-4.
Cui H, Zhang H, Ganger GR, Gibbons PB, **ng EP. Geeps: Scalable deep learning on distributed gpus with a gpu-specialized parameter server. In: Proceedings of the eleventh European conference on computer systems. EuroSys ’16. Association for computing machinery, New York, NY, USA 2016. https://doi.org/10.1145/2901318.2901323.
Haque A, Milstein A, Fei-Fei L. Illuminating the dark spaces of healthcare with ambient intelligence. Nature. 2020;585(7824):193–202. https://doi.org/10.1038/s41586-020-2669-y.
Kingma D, Ba J. Adam: A method for stochastic optimization. In: International conference on learning representations; 2014.
Sah R, Rodriguez-Morales A, Jha R, Chu D, Gu H, Peiris JS, Bastola A, Lal B, Ojha H, Rabaan A, Zambrano L, Costello A, Morita K, Pandey B, Poon L, Hopkins J, Healthcare A, Dhahran S. Arabia: Complete genome sequence of a 2019 novel coronavirus (sars-cov-2) strain isolated in Nepal. ASM Sci J. 2020. https://doi.org/10.1128/MRA.00169-20.
Lepikhin D, Lee H, Xu Y, Chen D, Firat O, Huang Y, Krikun M, Shazeer N, Chen Z. GShard: scaling giant models with conditional computation and automatic sharding; 2020. ar**v:2006.16668.
AI and compute. https://openai.com/blog/ai-and-compute. Accessed Jan 2021.
van der Maaten L, Hinton G. Viualizing data using t-sne. J Mach Learn Res. 2008;9:2579–605.
McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction; 2020. ar**v:1802.03426.
Benevolent AI. https://www.benevolent.com/. Accessed Jan 2021.
Chollet F. On the measure of intelligence; 2019. ar**v:1911.01547.
Nguyen TT. Artificial intelligence in the battle against coronavirus (COVID-19): a survey and future research directions; 2020. ar**v:2008.07343.
Raghu M, Schmidt E. A survey of deep learning for scientific discovery; 2020; ar**v:2003.11755.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A, Kaiser L, Polosukhin I. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:5998–6008.
Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in neural information processing systems, vol. 25. Curran Associates, Inc., 2012, p. 1097–1105. http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf. Accessed Jan 2021.
Gururangan S, Marasović A, Swayamdipta S, Lo K, Beltagy I, Downey D, Smith NA. Don’t stop pretraining: adapt language models to domains and tasks. In: ACL; 2020.
Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding.
Radford A. Improving language understanding by generative pre-training; 2018.
Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. Biobert: a pre-trained biomedical language representation model for biomedical text mining; 2019. https://doi.org/10.1093/bioinformatics/btz682. ar** a question answering dataset for COVID-19; 2020. ar**v:2004.11339.
Schick T, Schütze H. Exploiting cloze questions for few shot text classification and natural language inference; 2020. ar**v:2001.07676.
Schick T, Schütze H. It’s not just size that matters: small language models are also few-shot learners; 2020. ar**v:2009.07118.
Stebbing J, Phelan A, Griffin I, Tucker C, Oechsle O, Smith D, Richardson P. Covid-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis. 2020. https://doi.org/10.1016/S1473-3099(20)30132-8.
Wang Q, Huang L, Jiang Z, Knight K, Ji H, Bansal M, Luan Y. PaperRobot: incremental draft generation of scientific ideas.
Guarino N, Oberle D, Staab S. What is an ontology?. Berlin: Springer; 2009. p. 1–17. https://doi.org/10.1007/978-3-540-92673-3.
Wang Q, Li M, Wang X, Parulian N, Han G, Ma J, Tu J, Lin Y, Zhang H, Liu W, Chauhan A, Guan Y, Li B, Li R, Song X, Ji H, Han J, Chang S-F, Pustejovsky J, Rah J, Liem D, Elsayed A, Palmer M, Voss C, Schneider C, Onyshkevych B. COVID-19 literature knowledge graph construction and drug repurposing report generation; 2020. ar**v:2007.00576.
Davis AP, Grondin C, Johnson R, Sciaky D, King B, McMorran R, Wiegers J, Wiegers T, Mattingly C. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 2016;45:838. https://doi.org/10.1093/nar/gkw838.
Ilievski F, Garijo D, Chalupsky H, Divvala NT, Yao Y, Rogers C, Li R, Liu J, Singh A, Schwabe D, Szekely P. KGTK: a toolkit for large knowledge graph manipulation and analysis; 2020. ar**v:2006.00088.
Wise C, Ioannidis VN, Calvo MR, Song X, Price G, Kulkarni N, Brand R, Bhatia P, Karypis G. COVID-19 knowledge graph: accelerating information retrieval and discovery for scientific literature; 2020. ar**v:2007.12731.
Zeng X, Song X, Ma T, Pan X, Zhou Y, Hou Y, Zhang Z, Karypis G, Cheng F. Repurpose open data to discover therapeutics for COVID-19 using deep learning; 2020. ar**v:2005.10831.
Chen C, Ebeid IA, Bu Y, Ding Y. Coronavirus knowledge graph: a case study; 2020. ar**v:2007.10287.
Hill F, Cho K, Korhonen A. Learning distributed representations of sentences from unlabelled data. In: HLT-NAACL; 2016.
Hope T, Portenoy J, Vasan K, Borchardt J, Horvitz E, Weld DS, Hearst MA, West J. SciSight: Combining faceted navigation and research group detection for COVID-19 exploratory scientific search; 2020. ar**v:2005.12668.
WHO Infodemic Management. https://www.who.int/teams/risk-communication/infodemic-management. Accessed Jan 2021.
Alam F, Dalvi F, Shaar S, Durrani N, Mubarak H, Nikolov A, Martino GDS, Abdelali A, Sajjad H, Darwish K, Nakov P. Fighting the COVID-19 infodemic in social media: a holistic perspective and a call to arms; 2020. ar**v:2007.07996.
Dharawat A, Lourentzou I, Morales A, Zhai C. Drink bleach or do what now? Covid-HeRA: a dataset for risk-informed health decision making in the presence of COVID19 misinformation; 2020. ar**v:2010.08743.
Hossain T, Logan RL, Ugarte A, Matsubara Y, Young S, Singh S. Detecting COVID-19 misinformation on social media
Xu M, Ouyang L, Gao Y, Chen Y, Yu T, Li Q, Sun K, Bao F, Safarnejad L, Wen J, Jiang C, Chen T, Han L, Zhang H, Gao Y, Yu Z, Liu X, Yan T, Li H, Chen S. Accurately differentiating COVID-19, other viral infection, and healthy individuals using multimodal features via late fusion learning. https://doi.org/10.1101/2020.08.18.20176776.
Thorne J, Vlachos A, Christodoulopoulos C, Mittal A. Fever: a large-scale dataset for fact extraction and verification; 2018.
Wadden D, Lin S, Lo K, Wang LL, van Zuylen M, Cohan A, Hajishirzi H. Fact or fiction: verifying scientific claims; 2020. ar**v:2004.14974.
Heredia B, Prusa J, Khoshgoftaar TM. Exploring the effectiveness of twitter at polling the united states 2016 presidential election; 2017. p. 283–290. https://doi.org/10.1109/CIC.2017.00045.
Heredia B, Prusa J, Khoshgoftaar TM. Social media for polling and predicting united states election outcome. Soc Netw Anal Min. 2018. https://doi.org/10.1007/s13278-018-0525-y.
Nguyen DQ, Vu T, Rahimi A, Dao MH, Nguyen LT, Doan L. WNUT-2020 task 2: identification of informative COVID-19 english tweets; 2020. ar**v:2010.08232.
Chauhan K. NEU at WNUT-2020 ttask 2: data augmentation to tell bert that death is not necessarily informative; 2020. ar**v:2009.08590.
Sancheti A, Chawla K, Verma G. LynyrdSkynyrd at WNUT-2020 task 2: semi-supervised learning for identification of informative COVID-19 english tweets; 2020. ar**v:2009.03849.
Loon Av, Steward S, Waldon B, Lakshmikanth SK, Shah I, Guntuku SC, Sherman G, Zou J, Eichstaedt J. Not just semantics: social distancing and covid discourse on twitter.
Google Mobility Reports. https://www.google.com/covid19/mobility/. Accessed Jan 2021.
Castaneda G, Morris P, Khoshgoftaar TM. Investigation of maxout activations on convolutional neural networks for big data text sentiment analysis; 2019.
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, KÃpf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Chintala S. PyTorch: An imperative style, high-performance deep learning library.
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C. Corrado Gs, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Zheng X. Tensorflow: large-scale machine learning on heterogeneous distributed systems; 2016.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–52. https://doi.org/10.1007/s11263-015-0816-y.
Raghu M, Zhang C, Kleinberg J, Bengio S. Transfusion: understanding transfer learning for medical imaging; 2019. ar**v:1902.07208.
Topol E. Welcoming new guidelines for ai clinical research. Nat Med. 2020;26:1318–20. https://doi.org/10.1038/s41591-020-1042-x.
Rieke N, Hancox J, Li W, Milletari F, Roth H, Albarqouni S, Bakas S, Galtier MN, Landman B, Maier-Hein K, Ourselin S, Sheller M, Summers RM, Trask A, Xu D, Baust M, Cardoso MJ. The future of digital health with federated learning; 2020. ar**v:2003.08119.
Fredrikson M, Jha S, Ristenpart T. Model inversion attacks that exploit confidence information and basic countermeasures; 2015. p. 1322–1333. https://doi.org/10.1145/2810103.2813677.
Ryffel T, Trask A, Dahl M, Wagner B, Mancuso J, Rueckert D, Passerat-Palmbach J. A generic framework for privacy preserving deep learning; 2018. ar**v:1811.04017.
Johnson J, Khoshgoftaar TM. Survey on deep learning with class imbalance. J Big Data. 2019;6:27. https://doi.org/10.1186/s40537-019-0192-5.
Johnson J, Khoshgoftaar TM. Deep learning and data sampling with imbalanced big data; 2019. p. 175–183. https://doi.org/10.1109/IRI.2019.00038.
Leevy J, Khoshgoftaar TM, Bauder R, Seliya N. A survey on addressing high-class imbalance in big data. J Big Data. 2018. https://doi.org/10.1186/s40537-018-0151-6.
Farooq M, Hafeez A. COVID-ResNet: a deep learning framework for screening of COVID19 from radiographs; 2020. ar**v:2003.14395.
Wang S, Kang B, Ma J, Zeng X, **ao M, Guo J, Cai M, Yang J, Li Y, Meng X, Xu B. A deep learning algorithm using ct images to screen for corona virus disease (covid-19). MedRxiv. 2020. https://doi.org/10.1101/2020.02.14.20023028.
Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi A. COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images; 2020. ar**v:2004.02696.
He K, Fan H, Wu Y, **e S, Girshick R. momentum contrast for unsupervised visual representation learning; 2019. ar**v:1911.05722.
Chen T, Kornblith S, Norouzi M, Hinton G. A simple framework for contrastive learning of visual representations; 2020. ar**v:2002.05709.
Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6:1–48.
Zhang Y, Jiang H, Miura Y, Manning CD, Langlotz CP. Contrastive learning of medical visual representations from paired images and text; 2020. ar**v:2010.00747.
Sowrirajan H, Yang J, Ng AY, Rajpurkar P. MoCo pretraining improves representation and transferability of chest X-ray models; 2020. ar**v:2010.05352.
Huang G, Liu Z, Weinberger KQ. Densely connected convolutional networks. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR); 2017. p. 2261–2269.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR); 2016. p. 770–778.
Real E, Aggarwal A, Huang Y, Le QV. Regularized evolution for image classifier architecture search; 2019. ar**v:1802.01548.
Zoph B, Le QV. Neural architecture search with reinforcement learning; 2017. ar**v:1611.01578.
Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shen D, Shi Y. Lung infection quantification of COVID-19 in CT images with deep learning; 2020. ar**v:2003.04655.
Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E. Rapid AI development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis; 2020. ar**v:2003.05037.
Gopalan A, Juan D-C, Magalhaes CI, Ferng C-S, Heydon A, Lu C-T, Pham P, Yu G. Neural structured learning: training neural networks with structured signals. In: Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining. KDD ’20. Association for Computing Machinery, New York, NY, USA; 2020. p. 3501–3502. https://doi.org/10.1145/3394486.3406701.
Makary MA, Daniel M. Medical error–the third leading cause of death in the us. BMJ. 2016. https://doi.org/10.1136/bmj.i2139.
Frankle J, Carbin M. The lottery ticket hypothesis: finding sparse, trainable neural networks; 2018. ar**v:1803.03635.
Fan A, Stock P, Graham B, Grave E, Gribonval R, Jegou H, Joulin A. Training with quantization noise for extreme model compression; 2020. ar**v:2004.07320.
Wozniak: could a computer make a cup of coffee? https://www.fastcompany.com/1568187/wozniak-could-computer-make-cup-coffee. Accessed Jan 2021.
Barfoot T, Burgner-Kahrs J, Diller E, Garg A, Goldenberg A, Kelly J, Liu X, Naguib HE, Nejat G, Schoellig AP, Shkurti F, Siegel H, Sun Y, Waslander SL. Making sense of the robotized pandemic response: a comparison of global and canadian robot deployments and success factors; 2020. ar**v:2009.08577.
Murphy RR, Gandudi VBM, Adams J. Applications of robots for COVID-19 response; 2020. ar**v:2008.06976.
Dasari S, Ebert F, Tian S, Nair S, Bucher B, Schmeckpeper K, Singh S, Levine S, Finn C. RoboNet: large-scale multi-robot learning.
Srinivas A, Laskin M, Abbeel P. CURL: contrastive unsupervised representations for reinforcement learning; 2020. ar**v:2004.04136.
Wu Y, Yan W, Kurutach T, Pinto L, Abbeel P. Learning to manipulate deformable objects without demonstrations; 2019. ar**v:1910.13439.
Zhu H, Yu J, Gupta A, Shah D, Hartikainen K, Singh A, Kumar V, Levine S. The ingredients of real-world robotic reinforcement learning; 2020. ar**v:2004.12570.
Brinati D, Campagner A, Ferrari D, Locatelli M, Banfi G, Cabitza F. Detection of covid-19 infection from routine blood exams with machine learning: a feasibility study. MedRxiv. 2020. https://doi.org/10.1101/2020.04.22.20075143.
Collins FS, Morgan M, Patrinos A. The human genome project: Lessons from large-scale biology. Science. 2003;300(5617):286–90. https://doi.org/10.1126/science.1084564.
Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-amodio S, Strippoli P, Canaider S. An estimation of the number of cells in the human body. Ann Hum Biol. 2013. https://doi.org/10.3109/03014460.2013.807878.
Lopez-Rincon A, Tonda A, Mendoza-Maldonado L, Claassen E, Garssen J, Kraneveld A. Accurate identification of SARS-CoV-2 from viral genome sequences using deep learning. https://doi.org/10.1101/2020.03.13.990242.
Shiaelis N, Tometzki A, Peto L, McMahon A, Hepp C, Bickerton E, Favard C, Muriaux D, Andersson M, Oakley S, Vaughan A, Matthews PC, Stoesser N, Crook D, Kapanidis AN, Robb NC. Virus detection and identification in minutes using single-particle imaging and deep learning. MedRxiv. 2020. https://doi.org/10.1101/2020.10.13.20212035.
Mei X, Lee H-C, Diao K-Y, Huang M, Lin B, Liu C, **e Z, Ma Y, Robson P, Chung M, Bernheim A, Mani V, Calcagno C, Li K, Li S, Shan H, Lv J, Zhao T, **a J, Yang Y. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26:1–5. https://doi.org/10.1038/s41591-020-0931-3.
Zhou Y, Wang F, Tang J, Nussinov R, Cheng F. Artificial intelligence in covid-19 drug repurposing. Lancet Digital Health. 2020. https://doi.org/10.1016/S2589-7500(20)30192-8.
Herland M, Khoshgoftaar T, Wald R. A review of data mining using big data in health informatics. J Big Data. 2014;1:2. ar**v:2006.16668.
Senior A, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Zidek A, Nelson A, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones D, Silver D, Kavukcuoglu K, Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:1–5. https://doi.org/10.1038/s41586-019-1923-7.
Ahamed S, Samad M. Information Mining for COVID-19 research from a large volume of scientific literature; 2020. ar**v:2004.0208.
Rao R, Bhattacharya N, Thomas N, Duan Y, Chen X, Canny J, Abbeel P, Song YS. Evaluating protein transfer learning with TAPE; 2019. ar**v:1906:08230.
Gao W, Mahajan S, Sulam J, Gray J. Deep learning in protein structural modeling and design.
Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical assessment of methods of protein structure prediction (casp) âround xiii. Proteins Struct Funct Bioinform. 2019;87(12):1011–20. https://doi.org/10.1002/prot.25823.
Barabasi A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. https://doi.org/10.1038/nrg2918.
Hamilton WL, Ying R, Leskovec J. Representation learning on graphs: methods and applications; 2017. ar**v:1709.05584.
De Las Rivas J, Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol. 2010;6:1000807. https://doi.org/10.1371/journal.pcbi.1000807.
Zeroual A, Harrou F, Abdelkader D, Sun Y. Deep learning methods for forecasting covid-19 time-series data: a comparative study. Chaos Solitons Fractals. 2020;140:110121. https://doi.org/10.1016/j.chaos.2020.110121.
Kim M, Kang J, Kim D, Song H, Min H, Nam Y, Park D, Lee J-G. Hi-covidnet: Deep learning approach to predict inbound covid-19 patients and case study in South Korea; 2020. https://doi.org/10.1145/3394486.3412864.
Matthew Le, LSTLMN. Mark Ibrahim: neural relational autoregression for high-resolution COVID-19 forecasting.
Dandekar R, Barbastathis G. Quantifying the effect of quarantine control in Covid-19 infectious spread using machine learning. https://doi.org/10.1101/2020.04.03.20052084.
Arik SO, Li C-L, Yoon J, Sinha R, Epshteyn A, Le LT, Menon V, Singh S, Zhang L, Yoder N, Nikoltchev M, Sonthalia Y, Nakhost H, Kanal E, Pfister T. Interpretable sequence learning for COVID-19 forecasting; 2020. ar**v:2008.00646.
Meirom EA, Maron H, Mannor S, Chechik G. How to stop epidemics: controlling graph dynamics with reinforcement learning and graph neural networks; 2020. ar**v:2010.05313.
Clark K, Khandelwal U, Levy O, Manning CD. What does BERT look at? An analysis of BERT’s attention; 2019. ar**v:1906.04341.
Tang Y, Nguyen D, Ha D. Neuroevolution of self-interpretable agents; 2020. https://doi.org/10.1145/3377930.3389847. ar**v:2003.08165.
Mahendran A, Vedaldi A. Visualizing deep convolutional neural networks using natural pre-images. Int J Comput Vis. 2015. https://doi.org/10.1007/s11263-016-0911-8.
Yin H, Molchanov P, Li Z, Alvarez JM, Mallya A, Hoiem D, Jha N, Kautz J. Dreaming to distill: Data-free knowledge transfer via deepinversion. In: 2020 IEEE/CVF conference on computer vision and pattern recognition (CVPR); 2020. p. 8712–8721.
Hinton G, Vinyals O, Dean J. Distilling the knowledge in a neural network. In: NIPS deep learning and representation learning workshop; 2015. ar**v:1503.02531.
Ribeiro M, Singh S, Guestrin C. “why should i trust you?”: explaining the predictions of any classifier; 2016. p. 97–101. https://doi.org/10.18653/v1/N16-3020.
RAPIDS. https://www.rapids.ai/. Accessed Jan 2021.
Chan D, Rao R, Huang F, Canny J. t-SNE-CUDA: GPU-Accelerated t-SNE and its applications to modern data.
Narang S, Raffel C, Lee K, Roberts A, Fiedel N, Malkan K. WT5?! training text-to-text models to explain their predictions; 2020. ar**v:2004.14546.
Goodfellow IJ, Shlens J, Szegedy C. Explaining and harnessing adversarial examples; 2014. ar**v:1412.6572.
Winkens J, Bunel R, Roy AG, Stanforth R, Natarajan V, Ledsam JR, MacWilliams P, Kohli P, Karthikesalingam A, Kohl S, Cemgil T, Eslami SMA, Ronneberger O. Contrastive training for improved out-of-distribution detection; 2020. ar**v:2007.05566.
Ribeiro M, Wu T, Guestrin C, Singh S. Beyond accuracy: behavioral testing of nlp models with checklist; 2020. p. 4902–4912. https://doi.org/10.18653/v1/2020.acl-main.442.
Clark P, Tafjord O, Richardson K. Transformers as soft reasoners over language; 2020. ar**v:2002.05867.
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst. 2013;26:3111–9.
Kaplan J, McCandlish S, Henighan T, Brown TB, Chess B, Child R, Gray S, Radford A, Wu J, Amodei D. Scaling laws for neural language models; 2020. ar**v:2001.08361.
Finn C, Abbeel P, Levine S. Model-agnostic meta-learning for fast adaptation of deep networks; 2017.
Leevy J, Khoshgoftaar TM, Villanustre F. Survey on rnn and crf models for de-identification of medical free text. J Big Data. 2020;7:73. https://doi.org/10.1186/s40537-020-00351-4.
Hancock J, Khoshgoftaar TM. Survey on categorical data for neural networks. J Big Data. 2020. https://doi.org/10.1186/s40537-020-00305-w.
Bisk Y, Holtzman A, Thomason J, Andreas J, Bengio Y, Chai J, Lapata M, Lazaridou A, May J, Nisnevich A, Pinto N, Turian J. Experience grounds language; 2020. ar**v:2004.10151.
Tan H, Bansal M. Vokenization: improving language understanding with contextualized, visual-grounded supervision; 2020. ar**v:2010.06775.
Richter A, Khoshgoftaar T. A review of statistical and machine learning methods for modeling cancer risk using structured clinical data. Artif Intell Med. 2018. https://doi.org/10.1016/j.artmed.2018.06.002.
Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission; 2019. ar**v:1904.05342.
Johnson A, Pollard T, Shen L, Lehman L-W, Feng M, Ghassemi M, Moody B, Szolovits P, Celi L, Mark R. Mimic-iii, a freely accessible critical care database. Sci Data. 2016;3:160035. https://doi.org/10.1038/sdata.2016.35.
Acknowledgements
We would like to thank the reviewers in the Data Mining and Machine Learning Laboratory at Florida Atlantic University. Additionally, we acknowledge partial support by the NSF (IIS-2027890). Opinions, findings, conclusions, or recommendations in this paper are the authors’ and do not reflect the views of the NSF.
Funding
NSF RAPID (IIS-2027890).
Author information
Authors and Affiliations
Contributions
CS performed the literature review and drafted the manuscript. TMK worked with CS to develop the article’s framework and focus. TMK introduced this topic to CS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shorten, C., Khoshgoftaar, T.M. & Furht, B. Deep Learning applications for COVID-19. J Big Data 8, 18 (2021). https://doi.org/10.1186/s40537-020-00392-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40537-020-00392-9