Abstract
Background
The long introns of mammals are pools of evolutionary potential due to the multiplicity of sequences that permit the acquisition of novel exons. However, the permissibility of genes to this type of acquisition and its influence on the evolution of cell regulation is poorly understood.
Results
Here, we observe that human genes are highly permissive to the inclusion of novel exonic regions permitting the emergence of novel regulatory features. Our analysis reveals the potential for novel exon acquisition to occur in over 30% of evaluated human genes. Regulatory processes including the rate of splicing efficiency and RNA polymerase II (RNAPII) elongation control this process by modulating the “window of opportunity” for spliceosomal recognition. DNA damage alters this window promoting the inclusion of repeat-derived novel exons that reduce the ribosomal engagement of cell cycle genes. Finally, we demonstrate that the inclusion of novel exons is suppressed in hematological cancer samples and can be reversed by drugs modulating the rate of RNAPII elongation.
Conclusion
Our work demonstrates that the inclusion of repeat-associated novel intronic regions is a tightly controlled process capable of expanding the regulatory capacity of cells.
Similar content being viewed by others
Background
A major challenge in biology is to understand how complex regulatory networks emerge during evolution. An important mechanism for expanding complexity is alternative pre-mRNA splicing (AS), the process by which exonic regions are differentially excised to create multiple transcripts from a single gene locus. Recent surveys of organ transcriptomes across several vertebrate species have revealed AS profiles have diverged rapidly during vertebrate evolution, whereas organ mRNA expression profiles have remained relatively conserved [1, 2]. Moreover, several studies have described examples of how the emergence of lineage-specific isoforms can create novel phenotypes [3,4,5].
However, the emergence of AS-dependent complexity comes at a cost. On the one hand, AS confers flexibility to gene function by altering reading frame and tuning transcript stability [1, 4, 6,7,8]. On the other hand, the inappropriate recognition of intronic sequences resembling splice sites can give rise to the non-canonical execution of regulatory events disrupting gene expression [9]. These deleterious events often manifest themselves within human diseases [10]. Despite the importance of AS in evolution, the mechanisms and genomic features that control this balance between the promotion of novel functionality and its prevalence to cause disease are poorly understood.
In this study, we surveyed the human transcriptome to identify thousands of novel exonization events, the process by which non-canonical intronic sequences are incorporated into mRNA transcripts. We reveal these events do not occur randomly within the genome but are enriched within cell cycle and cell signaling genes. Exonization events occur within m6a (N6-methyladenosine)-modified long introns close to the transcription start site and often overlap Alu and L1 transposon events. The inclusion of these novel exons is promoted by regulatory events that promote the “window of opportunity” for spliceosome recognition, such as the rate of RNA polymerase II elongation and splicing efficiency dynamics. This multilayered system can be actively regulated by exogenous agents permitting the emergence of novel regulation as exemplified by UV irradiation, which promotes exonization within cell cycle genes to suppress their ribosomal engagement. We further provide evidence exonization is suppressed in hematological cancers. Thus, we identify a highly evolvable mechanism that can expand the regulatory complexity of cells.
Results
Exonization events occur in introns enriched with new transposons
To investigate the potential for novel exonization events to occur within the human transcriptome, we analyzed over 400 shRNA knockdown RNA-seq datasets from HepG2 cell lines [11] to identify reads map** between known exons and novel intronic sequences (Fig. 1a and the “Methods” section). We only considered reads map** to exon-exon junctions (EEJs) supported by at least 5 reads and a percent spliced in (PSI) value of at least 5%. Novel exons were defined as those absent from annotation databases [13, 14] and all non-perturbed control datasets (Fig. 1a). Confirming the validity of this approach, the knockdown of the RNA binding protein (RBP) heterogeneous ribonucleoprotein C (hnRNPC) created the most Alu-derived exonization events (Additional file 1: Figure S1), in line with previous observations [15]. In total, we detected 13,103 novel exonic events within 4774 genes or 30.6% of evaluated human protein-coding genes under the perturbations we surveyed.
Genomics features of introns with exonization events. a Workflow to identify novel exonization events (see the “Methods” section). Briefly, RNA-seq from shRNA knockdown of RNA binding proteins in HepG2 is analyzed by 2-pass enabled STAR, and then novel junctions are incorporated into index files analyzed by Whippet [12]. Identified exons are filtered to remove exon-exon junctions and events occurring in any of the matched control samples, as well as annotated in genome databases. Only events supported by > 5 reads map** over exon-exon junctions and a percent spliced in (PSI) greater than 5% are included. b Plot showing the results from a logistic linear regression analysis aimed at identifying features important in discriminating introns prone to exonization events to all other expressed introns. Features in bold significantly contribute to the model (p < 0.01, Student t test). TSS, transcription start site; ppt_len, polypyrimidine tract length; 5′ss, 5′-splice site; 3’ss, 3′-splice site; bp_scr, branchpoint score; SS_dist, splice site distance; BP_num, branchpoint number; AGEZ, AG dinucleotide Exclusion Zone length; TAD, topologically associating domain; ppt_scr, polypyrimidine tract score (n = 13,103). c Plot showing the results from a logistic linear regression analysis aimed at identifying the type of transposable elements that most effectively discriminate introns prone to exonization events compared to all other expressed introns. Features in bold significantly contribute to the model (p < 0.01, Student t test). Nodes are colored by average estimated age of when transposable elements arose (n = 13,103). d Enrichment map for GO, REACTOME, and KEGG functional categories of genes that contain Alu-exonization events, with representative GO terms shown for each sub-network (see Additional file 1: Figure S1 for annotated version). Node size is proportional to the number of genes associated with the GO category, and edge width is proportional to the number of genes shared between GO categories
To investigate the mechanisms underlying these exonization events, we collated a list of features classically associated with alternative splicing, including splice site strength, GC content, and polypyrimidine tract length (Additional file 2: Table S1). Logistic linear regression was then used to compare these events with a “background” group of expressed introns lacking any evidence of exonization (Fig. 1b). Validating our choice of genomic features, our model achieves a high average true positive rate [AUC, area under the receiver operating characteristic (ROC) curve] of 75.2% (Additional file 1: Figure S1). Moreover, we were able to confirm previous results that exonization events tend to occur in long introns with a high GC content [16] (Additional file 1: Figure S1, intron length: p < 3.53 × 10−59, GC 1.01 × 10−73, Student t test). Notably, we find exonization events often overlap nucleosome-binding sites (Additional file 1: Figure S1, p < 2.37 × 10−23, Wilcoxon test), rarely occur at the 3´-end of the gene body (p < 5.90 × 10−21, Student t test) and show a significant tendency to occur within 5´-UTRs (Additional file 1: Figure S1, p < 7.13 × 10−166, Fisher exact test). Importantly, we also observe that the strongest predictor for exonization was the occurrence of transposable elements overlap** the novel exon (Fig. 1b and Additional file 1: Figure S1, p < 6.46 × 10−127, Student t test). In comparison to these strong predictors, no cis-regulatory splicing elements contribute significantly to the model or show significant differences between the datasets (Additional file 1: Figure S1, p > 0.05, Student t test).
To evaluate the conservation of novel exon usage across species, we analyzed the extent of exonization across multiple matched tissue types within four primate species spanning 30 million years of primate evolution. To explore exonization usage between samples, genes with events occurring in all four species were identified and sorted using affinity propagation clustering. In line with canonical alternative splicing [1, 2], samples from the same species invariably clustered together (Additional file 1: Figure S2). The notable exception to this trend was observed in samples from the testes, which showed tissue-specific clustering. This suggests within the testes there is a strong exonization signature conserved across primate species in multiple genes (Additional file 1: Figure S2).
To further investigate the influence of retrotransposons on exonization, we sub-divided the transposable elements into their major sub-families. In line with the lineage-specificity of exonization events, the most significant contributors to the model are transposons younger than 70 million years. In particular, Alu element sub-family members AluJ and AluS, as well as the highly mobile L1 elements are strong predictors (Fig. 1c). Interestingly, an exception to this rule is the AluY sub-family [17], which shows a pattern reminiscent of much older transposon events (Fig. 1c). This difference is potentially due to its relative depletion within gene bodies (3%, 19%, AluY, AluS, occurrence in expressed introns). Finally, we wished to assess which type of genes contains Alu-exonization events. Interestingly, we find a strong enrichment for functions related to cell signaling and cell cycle regulation (Fig. 1d, Additional file 1: Figure S1 and Additional file 3: Table S2). Alongside previous examples [8, 9, 15, 18], our observation of an extensive number of exonization events overlap** new transposons suggests a novel source of transcriptomic complexity.
m6a RNA binding proteins suppress exonization
An evaluation of the trans-factors promoting exonization (Additional file 1: Figure S2 and Additional file 4: Table S3) revealed an enrichment of m6a (N6-methyladenosine) binding RBPs, especially among Alu-containing novel exons (Fig. 2a, p < 0.05, hypermetric test). This included hnRNPC, which has been previously shown to induce a large number of Alu-specific exonization events [15], as well as DiGeorge syndrome critical region 8 (DGCR8) and YTH domain-containing protein 2 (YTHDC2). To examine the potential role of m6a marks in exonization, we analyzed knockdown data of the m6a modification enzyme N6-adenosine-methyltransferase subunit (METTL3) [18]. This analysis revealed a significant increase in the number of detectable exonization events upon METTL3 knockdown (Fig. 2b, p < 3.57 × 10−03, Wilcox-rank sum test), in concordance m6a regulating the inclusion of novel exons. Further analysis of these METTL3-dependent exonization events revealed a functional enrichment of genes associated with DNA damage (p < 2.68 × 10−02, FDR-corrected p value). Next, we analyzed data from HeLa cells constituting of two knockdowns of known m6a regulators (Serine/arginine-rich splicing factor 3 (SRSF3) and YTH domain-containing protein 1 (YTHDC1)) and two knockdowns of RBPs not known to directly recognize m6a (Serine/arginine-rich splicing factor 9 and 10 (SRSF9 and SRSF10)) [19, 2c, p < 3.23 × 10−123, Wilcoxon rank-sum test; Fig. 2d, p < 2.13 × 10−63, Wilcoxon rank sum test; Additional file 1: Figure S2). Altogether, this suggests m6a modification and binding proteins are key regulators of (Alu-containing) exonization events.
Mechanisms increasing the “window of opportunity” for spliceosome recruitment promote exonization
Our initial analysis revealed the prediction of exonization is strongly enhanced by repeat elements, intron length and GC content. These features are known to negatively correlate with RNA polymerase II (RNAPII) elongation rate [23]. We therefore hypothesized that changes in RNAPII elongation rate may promote exonization. To test this, we analyzed RNA-seq data from human cells expressing mutations that increase (E1126G) or decrease (R749H) the elongation rate of RNA polymerase II (RNAPII) [24]. To determine the elongation rate for each mutant, we analyzed data from genome-wide nuclear run-on sequencing (GRO-seq) assay combined with transcription elongation inhibitor DRB (see the “Methods” section and [24]). We observe that mutations slowing the rate of RNAPII elongation (R749H) strongly induced exonization events (Fig. 3a, ALL: p < 3.18 × 10−45, Fisher’s exact test), with this change especially strong in Alu-containing novel exons (Fig. 3a, ALU: p < 5.62 × 10−53, Fisher’s exact test). In contrast, mutations that speed up elongation had negligible effects on the number of exonization events detected (Fig. 3a; p > 0.05, Fisher’s exact test).
Reduced rate of RNA polymerase II elongation and poor splicing efficiency promotes exonization. a Dot plot showing the impact of RNA polymerase mutations on exonization of Alu-containing exons. WT, wildtype; Fast, E1126G mutation; Slow, R749H mutation. Each point represents individual dataset. KB, kilobase; min, minutes. Elongation, rate of transcriptional elongation (see the “Methods” section and [24]). b Boxplot of splicing efficiency for introns with exonization events vs all expressed introns with no evidence of exonization. Splicing efficiency is a metric describing speed of intron excision as measured by assessing nascent RNA-seq using BrU-chase at 0, 15, 30, and 60 min. See Fig. 2d for the description of boxplots. ***p < 1 × 10−10 p value calculated using Wilcoxon-rank sum test. (n = 4,011). c Stacked bar plot showing distributions of introns for splicing efficiency identified by BrU-chase. Groups assigned by K-means clustering (k = 5) (see the “Methods” section)—see Additional file 1: Figure S3a for distributions of splicing efficiencies. (n = 83,972)
This result supports the competition model of alternative splicing [24, 25] wherein the regulation of exon inclusion is associated a “window of opportunity” for spliceosome recognition. If this connection between exonization and opportunity is valid, an independent mechanism for the emergence of novel exons should occur within introns that are slowly processed by the splicing machinery. To evaluate this hypothesis, we analyzed nascent RNA data from BrU-Chase-seq [21], in which cells are labeled with a 15-min BrU pulse and chased for 0, 15, 30, and 60 min. To determine splicing kinetics for each intron, we calculated the splicing efficiency dynamics (SEDs) or the rate of intron excision [21, 26] (see the “Methods” section). K-means clustering was then used to identify five groups of introns with SEDs ranging from very fast to very slow (Additional file 1: Figure S3). Introns containing exonization events were then compared to a background set of all expressed introns. Strikingly, this analysis reveals that introns containing exonization events are strongly enriched within the slowest SED cluster (Fig. 3b, p < 3.23 × 10−239, Wilcox rank-sum test, and Additional file 1: Figure S3). Moreover, these introns display a highly significant reduction in SED compared to background groups (Fig. 3c and Additional file 1: Figure S3, p < 2.2 × 10−160, Wilcoxon rank-sum test). Together, these observations suggest that mechanisms that expand the “window of opportunity” will increase the likelihood of recognition by the splicing machinery and thereby promote the rate of exonization.
DNA damage induces exonization within cell cycle genes
Exogenous process can also promote alterations in transcription elongation and therefore may alter rates of exonization. To investigate this we focused on UV irradiation as previous studies have demonstrated that it promotes both the hyperphosphorylation of RNAPII leading to the subsequent inhibition of transcription elongation [27, 28] and the recruitment of the m6a machinery to sites of DNA damage [25, 38] for the spliceosome to recognize novel exons. We highlight this can occur by decreasing the rate of RNAPII elongation and is associated with slow splicing efficiency dynamics. These novel exons are also marked by m6a RNA modifications. This multi-layered system permits exogenous forces to regulate exonization (Fig. 6). We demonstrate UV irradiation increases the rate of exonization within cell cycle genes, potentially by slowing RNAPII elongation [27, 28], and observe that exonization within these genes coincides with reduced polysome engagement. Furthermore, we describe in cancer how this “window of opportunity” mechanism is repressed and link this suppression to particular cancer mutations [34] within RNA binding splicing factors. Collectively, these results provide new insights into the control and dynamics of exonization in different biological and disease contexts, as well as highlighting an evolutionary process with the potential to expand regulatory complexity within cells.
A model summarizing results from this paper. A model summarizing results from this paper contrasting regulatory mechanisms associated with opening (or facilitating) and closing (or inhibiting) the window of opportunity for exonization. RBP, RNA binding protein; RNAP II, RNA polymerase II; m6a, N6-methyladenosine
Previous work has shown that RBPs and nucleosome occupancy underlie the regulatory control of exonization. For example, competition between the hnRNPC and the 3′-splicing factor U2AF2 [18], in tandem with nonsense-mediated decay [15], has been shown to restrict the inclusion of Alu-containing exons. On the other hand, high nucleosome occupancy is associated with the emergence of new exons [7, 16] and proposed to promote RNA polymerase II pausing [7, 16, 39]. Our observations expand these findings identifying a “window of opportunity” model [25, 38] for exonization controlled by a multi-layered regulatory program, including m6A-associated RBPs that suppress the emergence of new exons. The regulatory networks controlling exonization events are highly interconnected, as RNAPII facilitates the deposition of m6a onto actively transcribing nascent transcripts [40], which is known to tune splicing efficiency [21]. Given our observations that exonization is subject to multi-layered regulatory control (i.e., RNAP-II, RBPs and m6A—see Fig. 6), it is also interesting to consider how this mechanism may influence the life cycle of a transcript. Our results show exonization is associated with decreases in polysome association of genes containing exonization events. An explanation for this observation is that short sequences derived from Alu elements [30] and transcripts with other repeats [41, 42] have increased nuclear accumulation, which would restrict the ribosomal accessibility of transcripts with exonization events (Additional file 1: Figure S4).
It also noteworthy that a system likely evolved to suppress the aberrant impact of transposon inclusion on functional transcripts [43] may have been co-opted to create a novel regulatory mechanism. In support of this proposal, we identify UV irradiation is accompanied by an increase in exonization within cell cycle genes potentially restricting the expression of key checkpoint regulators, until the DNA damage process is complete. These changes may be the result of perturbations to the multi-layered regulatory network controlling exonization, for example, UV irradiation slows the elongation rate of RNA polymerase II [28], and DNA damage up-regulates the m6a-regulatory machinery [48]. Further quality checks were done using Trimmomatics [49] to remove adaptors, low-quality reads, and all reads less than 50 nucleotides in length.
Datasets
All datasets used described in Additional file 5: Table S4 [2, 11, 21, 24, 28, 31, 50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65].
Alternative splicing RNA-seq analysis
Whippet [12] was used to analyze RNA-Seq data employed for the identification of exonization events. Whippet quantifies all combinations of EEJs, including cassette, mutually exclusive, and microexon events. Whippet (v1.0) was run using default settings with “—biascorrect” option enacted to correct for 5´-sequence and GC content batch/bias errors (https://github.com/timbitz/Whippet.jl).
To create the splice graphs required for Whippet splicing quantification, genome annotation files were extracted from Ensembl (Hg38 – Release 93) [69]. For each dataset, this was supplemented with novel exon-exon junctions derived from whole-genome alignment by STAR [66] with 2-pass setting enabled and outFilterMultimapNmax == 10. Whippet index was run for each dataset with “—bam” setting enabled and “--suppress-low-tsl.” Whippet quantification using bias correction function enabled to correct 5′ sequence and GC content bias. Otherwise, the default settings were used, so that only reads map** to exon-exon junctions were used to quantify splicing. Bedtools (intersect –v) [67] was used to remove all exons overlap** with annotation from UCSC genome browser [13]. UCSC Liftover [13] was used to convert all non-human exons in conservation analysis.
Gene expression RNA-seq analysis
Kallisto [68] was used with default settings with index constructed using data extracted from Ensembl Hg38 Release 93 [69].
Identification of exonization events
All events identified by Whippet were only considered as novel exonization events if they passed the following criteria: (1) Exon inclusion was supported by at least 5 corrected reads, as assigned by expectation maximization by Whippet. (2) Event must be CE (core exon) event. (3) Exon was not identified in any of the control/matched datasets. (4) Exon was not previously annotated in Ensembl Hg38 Gene transfer format (GTF) file or UCSC GTF file. (5) Exon had a percent-spliced in (PSI) value of at least 0.05 (i.e., 5%). (6) Exon-exon junction reads must occur between novel exon and known exon.
An exon was considered previously annotated if either exon-exon junction was annotated in Hg38 GTF file (from Ensembl or UCSC). The “number of exonization event” are all those events identified in this manner.
Overlap with known repeat elements
Repeat elements identified by RepeatMasker were downloaded from UCSC table browser [13] in bed format. Bedtools intersect (−wao –f 0.2) was used to identify overlap of transposons with novel exons.
Frequency of Alu-transposable events is calculated as the proportion of exonization events overlap** transposons that are identified as Alu events. All Alu events identified by repeatmasker (containing annotation “Alu”) were grouped together.
Visualization of events
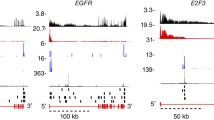
Visualization of splicing events were done using –-bam setting for whippet quant and visualized with Sashimi plots in IGV browser (−DenableSashimi = “true”) [70].
Functional analysis
Functional enrichment analysis was performed using the g:Profiler (https://biit.cs.ut.ee/gprofiler/gost) tool [71]. Genes identified as containing novel splicing events were compared to a background of genes expressed in sample (cRPKM or TPM > 1). Structured controlled vocabularies from Gene Ontology organization, as well as information from the curated KEGG and Reactome databases were included in the analysis. Only functional categorizes with more than five members and fewer than 2000 members were included in the analysis. Significance was assessed using the hypergeometric test. p values were corrected for multiple testing using the method of Benjamini-Hochberg. The Cytoscape application EnrichmentMap (baderlab.org/Software/EnrichmentMap) was used to visualize functional enrichment [72].
General logistic regression
All continuous data was normalized to ensure fair comparison between features. The R module GLM with default setting except family = binomial(). Data was split into training and test data with 90:10% split. ROC curve calculated using test data using ROCR library.
Exonic features
MaxEntScan [73] was used to estimate the strength of 3′ and 5′ splice sites. 5′ splice site strength was assessed using a sequence including 3 nt of the exon and 6 nt of the adjacent intron. 3′ splice site strength was assessed using a sequence including − 20 nt of the flanking intron and 3 nt of the exon. SVM-BPfinder [74] was used to estimate branchpoint and polyprimidine tract strength and other statistics. Score was estimated using the sequence of introns to the 3’end of exon between 20 and 500 nt.
Transcription start sites (TSS) were downloaded from Biomart. TAD boundaries for HepG2 were extracted from ENCODE [11] pre-processed data and converted to Hg38 by liftover. GC content was calculated using python script. Transposon information download from RepeatMasker as described above.
Nucleosome occupancy for HepG2 cells was calculated using data from Enroth et al. [75]. Colorspace read data was aligned using Bowtie [76] (-S -C -p 4 -m 3 --best –strata) using index file constructed from Ensembl Hg38. Nuctools (with default settings) was used to calculate occupancy profiles and calculate occupancy at individual regions [77].
In feature analysis, only exonization events within introns detected in this analysis were used.
Splicing efficiency dynamics
Splicing efficiency dynamics was calculated using approach described previously [26]. Briefly, reads were mapped to Ensembl Hg38 assembly using STAR (2-pass enabled) and only uniquely mapped reads kept for downstream analysis. Splicing index values were first calculated which represent ratio of the split reads map** to the 5′ and 3′ SJ of an intron divided to the sum of split plus non-split reads. The θ value (representing Splicing Efficiency, SE) was extracted from all pulse-chase time points, for introns with at least five reads coverage at both 5′ and 3′ SJ. K-means clustering used to identify give groups of distinct splicing efficiency (very fast, fast, medium, slow and very slow). The Splicing Efficiency Dynamics metric was calculated as SED = 1/ ((1.001 − θ 0 min) × (1.001 − θ 60 min)).
Identification of m6a reads
After genome-wide map** to Hg38 assembly using STAR (2-pass enabled) [66], CLAM (CLiP-seq Analysis of Multi-mapped reads) [Nascent RNA-seq analysis (including GRO-seq) Wavefront and elongation speeds were extracted from supplementary data of relevant papers [23, 28]. Reads were randomly sampled from a 100 M single-end HeLa RNA-seq dataset using the program “fastq-sample” from the “fastq-tools” (v0.8) pipeline using randomized seeds and no replacement. Identical pipeline (see “Identification of exonization events” section of methods) was run on every dataset and the percentage of Alu elements detected.Investigation of influence of read depth on detection of novel exons
Availability of data and materials
All packages and publicly available computer code are described in the “Methods” section of this manuscript. Whippet is freely available at https://github.com/timbitz/Whippet.jl [12]. All plots were created using R Package ggplot and scripts are available upon requests. All data generated during this study are included in this article and its supplementary files or are available from external sources: all datasets analyzed in this study are available in SRA (list of identifiers and references provided in Additional file 5: Table S4 [2, 11, 21, 24, 28, 31, 50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65]). Additional in-house scripts with associated data-files are available in the Figshare repository [78].
References
Barbosa-Morais NL, Irimia M, Pan Q, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–93.
Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338:1593–9.
Gracheva EO, Cordero-Morales JF, González-Carcacía JA, et al. Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature. 2011;476:88–91.
Gueroussov S, Weatheritt RJ, O’Hanlon D, et al. Regulatory expansion in mammals of multivalent hnRNP assemblies that globally control alternative splicing. Cell. 2017;170:324–39 e23.
Nicolas M, Rodríguez-Buey ML, Franco-Zorrilla JM, Cubas P. A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr Biol. 2015;25:1799–809.
Gueroussov S, Gonatopoulos-Pournatzis T, Irimia M, et al. An alternative splicing event amplifies evolutionary differences between vertebrates. Science. 2015;349:868–73.
Merkin JJ, Chen P, Alexis MS, Hautaniemi SK, Burge CB. Origins and impacts of new mammalian exons. Cell Rep. 2015;10:1992–2005.
Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced.[letter]. Genome Res. 2002;12(7):1060–7.
Attig J, Agostini F, Gooding C, et al. Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell. 2018;174:1067–81 e17.
Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32.
Sundararaman B, Zhan L, Blue SM, et al. Resources for the comprehensive discovery of functional RNA elements. Mol Cell. 2016;61:903–13.
Sterne-Weiler T, Weatheritt RJ, Best AJ, Ha KCH, Blencowe BJ. Efficient and accurate quantitative profiling of alternative splicing patterns of any complexity on a laptop. Mol Cell. 2018;72(1):187–200.e6.
Haeussler M, Zweig AS, Tyner C, et al. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2018.
Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–61.
Zarnack K, König J, Tajnik M, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–66.
Li Y, Li C, Li S, et al. Human exonization through differential nucleosome occupancy. Proc Natl Acad Sci U S A. 2018;115:8817–22.
Hellen EH, Brookfield JF. Alu elements in primates are preferentially lost from areas of high GC content. PeerJ. 2013;1:e78.
Attig J, Ruiz de Los Mozos I, Haberman N et al. Splicing repression allows the gradual emergence of new Alu-exons in primate evolution. Elife. 2016;5:e19545.
Kasowitz SD, Ma J, Anderson SJ, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412.
**ao W, Adhikari S, Dahal U, et al. Nuclear m (6) a reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–19.
Louloupi A, Ntini E, Conrad T, Ørom UAV. Transient N-6-methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 2018;23:3429–37.
Zhang Z, **ng Y. CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome. Nucleic Acids Res. 2017;45:9260–71.
Veloso A, Kirkconnell KS, Magnuson B, et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24:896–905.
Fong N, Kim H, Zhou Y, et al. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28:2663–76.
Dujardin G, Lafaille C, de la Mata M, et al. How slow RNA polymerase II elongation favors alternative exon skip**. Mol Cell. 2014;54:683–90.
Mukherjee N, Calviello L, Hirsekorn A, de Pretis S, Pelizzola M, Ohler U. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol. 2017;24:86–96.
Muñoz MJ, Pérez Santangelo MS, Paronetto MP, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–20.
Williamson L, Saponaro M, Boeing S, et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell. 2017;168:843–55 e13.
**ang Y, Laurent B, Hsu CH, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–6.
Lubelsky Y, Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555:107–11.
Wahba A, Rath BH, Bisht K, Camphausen K, Tofilon PJ. Polysome profiling links translational control to the radioresponse of glioblastoma stem-like cells. Cancer Res. 2016;76:3078–87.
Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int J Cell Biol. 2013;2013:463786.
Sebestyén E, Singh B, Miñana B, et al. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016;26:732–44.
Ilagan JO, Ramakrishnan A, Hayes B, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25:14–26.
Okeyo-Owuor T, White BS, Chatrikhi R, et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29:909–17.
Shirai CL, Ley JN, White BS, et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27:631–43.
Patterson SE, Liu R, Statz CM, Durkin D, Lakshminarayana A, Mockus SM. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum Genomics. 2016;10:4.
Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–75.
Gilchrist DA, Dos Santos G, Fargo DC, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–51.
Huang H, Weng H, Zhou K, et al. Histone H3 trimethylation at lysine 36 guides m. Nature. 2019;567:414–9.
Carlevaro-Fita J, Das M, Polidori T, Navarro C, Johnson R. Ancient exapted transposable elements promote nuclear enrichment of long noncoding RNAs. biorxiv. 2018; https://doi.org/10.1101/189753
Fazal FM, Han S, Kaewsapsak P, et al. Atlas of Subcellular RNA Localization Revealed by APEX-seq. biorxiv. 2018. https://doi.org/10.1101/454470.
Sibley CR, Blazquez L, Ule J. Lessons from non-canonical splicing. Nat Rev Genet. 2016;17:407–21.
Anantha RW, Alcivar AL, Ma J, et al. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS One. 2013;8:e61368.
McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–28.
Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m. Nature. 2017;552:126–31.
Laumont CM, Vincent K, Hesnard L, et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med. 2018;10(470).
Wingett SW, Andrews S. FastQ Screen: A tool for multi-genome map** and quality control. F1000Res. 2018;7:1338.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Saenz DT, Fiskus W, Qian Y, et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia. 2017;31:1951–61.
Brawand D, Soumillon M, Necsulea A, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–8.
Cavga AD, Tardu M, Korkmaz T, et al. Cryptochrome deletion in p53 mutant mice enhances apoptotic and anti-tumorigenic responses to UV damage at the transcriptome level. Funct Integr Genomics. 2019. doi: https://doi.org/10.1007/s10142-019-00680-5.
Chen L, Chen JY, Huang YJ, et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol Cell. 2018;69:412–25 e6.
Collier AE, Spandau DF, Wek RC. Translational control of a human. Mol Biol Cell. 2018;29:29–41.
Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. Elife. 2016;5:1092.
Jiang L, Huang J, Higgs BW, et al. Genomic landscape survey identifies SRSF1 as a key Oncodriver in small cell lung Cancer. PLoS Genet. 2016;12:e1005895.
Lee SH, Singh I, Tisdale S, Abdel-Wahab O, Leslie CS, Mayr C. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018;561:127–31.
Louloupi A, Ntini E, Liz J, Ørom UA. Microprocessor dynamics shows co- and post-transcriptional processing of pri-miRNAs. RNA. 2017;23:892–8.
Pellagatti A, Armstrong RN, Steeples V, et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood. 2018;132:1225–40.
Sanlorenzo M, Vujic I, Esteve-Puig R, et al. The lincRNA MIRAT binds to IQGAP1 and modulates the MAPK pathway in NRAS mutant melanoma. Sci Rep. 2018;8:10902.
Siebenthall KT, Miller CP, Vierstra JD, et al. Integrated epigenomic profiling reveals endogenous retrovirus reactivation in renal cell carcinoma. EBioMedicine. 2019;41:427–42.
Stevens BM, Khan N, D’Alessandro A, et al. Characterization and targeting of malignant stem cells in patients with advanced myelodysplastic syndromes. Nat Commun. 2018;9:3694.
Suntsova M, Gaifullin N, Allina D, et al. Atlas of RNA sequencing profiles for normal human tissues. Sci Data. 2019;6:36.
Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19:A68–77.
Wenric S, ElGuendi S, Caberg JH, et al. Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Sci Rep. 2017;7:17452.
Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Quinlan AR. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11 12.1–34.
Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7.
Cunningham F, Achuthan P, Akanni W, et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–51.
Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92.
Reimand J, Arak T, Adler P, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016;44:W83–9.
Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482–517.
Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94.
Corvelo A, Hallegger M, Smith CW, Eyras E. Genome-wide association between branch point properties and alternative splicing. PLoS Comput Biol. 2010;6:e1001016.
Enroth S, Andersson R, Bysani M, et al. Nucleosome regulatory dynamics in response to TGFβ. Nucleic Acids Res. 2014;42:6921–34.
Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010;Chapter 11:Unit 11.7.
Vainshtein Y, Rippe K, Teif VB. NucTools: analysis of chromatin feature occupancy profiles from high-throughput sequencing data. BMC Genomics. 2017;18:158.
Weatheritt RJ, Avgan N, Wang JI, Fernandez-Chamorro J. Multilayered control of exon acquisition permits the emergence of novel forms of regulatory control. 2019 Figshare https://doi.org/10.6084/m9.figshare.8797067.
Acknowledgements
We gratefully acknowledge B. Harper, K. Ha, S. Chavali, and S. Gueroussov for helpful discussions and critical review of the manuscript.
Review history
The review history is available as Additional file 7.
Funding
This work was supported by an EMBL Australia Fellowship to R.J.W. R.J.W. is a Scrimshaw Fellow.
Author information
Authors and Affiliations
Contributions
NA, JIW, JFC, and RJW designed and undertook the experiments. RJW wrote the manuscript and designed the project with input from other authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Extension of Fig. 1 displaying the genomic features of introns associated with exonization events. Figure S2. Tissue-specific and shRNA analysis of novel exonic events. Figure S3. Extension of Fig. 3 displaying the analysis of the splicing efficiency dynamics. Figure S4. Extension of Fig. 4 showing evaluation of the UV exposure data in human and mouse samples. Figure S5. Extension of Fig. 5, and control analysis for detection of repeat elements at differing gene-expression levels and read depths. (PDF 2584 kb)
Additional file 2:
Table S1. Genomic and splicing features used in logistic linear regression model. (XLSX 40 kb)
Additional file 3:
Table S2. GO terms results for genes with exonization events. (XLSX 109 kb)
Additional file 4
Table S3. Coordinates, gene names and associated RNA binding knockdown identification for identified novel exon events.) (XLSX 764 kb)
Additional file 5:
Table S4. Datasets used in paper including description and relevant figures information plus PubMed ID and SRA ID. (XLSX 54 kb)
Additional file 6:
Table S5. GO terms results from GProfiler for exonization events identified depleted in cancer samples. (XLSX 254 kb)
Additional file 7:
Review history. (DOCX 30 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Avgan, N., Wang, J.I., Fernandez-Chamorro, J. et al. Multilayered control of exon acquisition permits the emergence of novel forms of regulatory control. Genome Biol 20, 141 (2019). https://doi.org/10.1186/s13059-019-1757-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13059-019-1757-5