Abstract
Background
Kidney transplantation is the optimal treatment to cure the patients with end-stage renal disease (ESRD). However, the infectious complication, especially pneumonia, is the main cause of mortality in the early stage. Immune monitoring by relevant biomarkers provides direct evidence of immune status. We aimed to study the association between immune monitoring and pneumonia in kidney transplant patients through machine learning models.
Methods
A total of 146 patients receiving the immune monitoring panel in our center, including 46 pneumonia recipients and 100 stable recipients, were retrospectively reviewed to develop the models. All the models were validated by external data containing 10 pneumonia recipients and 32 stable recipients. The immune monitoring panel consisted of the percentages and absolute cell counts of CD3+CD4+ T cells, CD3+CD8+ T cells, CD19+ B cells and natural killer (NK) cells, and median fluorescence intensity (MFI) of human leukocyte antigen (HLA)-DR on monocytes and CD64 on neutrophils. The machine learning models including support vector machine (SVM), logistic regression (LR), multi-layer perceptron (MLP) and random forest (RF) were applied for analysis.
Results
The pneumonia and stable groups showed significant difference in cell counts of each subpopulation and MFI of monocyte HLA-DR and neutrophil CD64. The SVM model by monocyte HLA-DR (MFI), neutrophil CD64 (MFI), CD8+ T cells (cells/μl), NK cells (cell/μl) and TBNK (T cells, B cells and NK cells, cells/μl) had the best performance with the average area under the curve (AUC) of 0.940. The RF model best predicted the patients who would progress into severe pneumonia, with the average AUC of 0.760. All the models had good performance validated by external data.
Conclusions
The immune monitoring panel was tightly associated with pneumonia in kidney transplant recipients. The models developed by machine learning techniques identified patients at risk and predicted the prognosis. Based on the results of immune monitoring, better individualized therapy might be achieved.
Similar content being viewed by others
Background
Kidney transplantation is the optimal treatment to cure the patients with end-stage renal disease (ESRD) [1]. Novel immunosuppressive drugs improve the prognosis of kidney transplantation and minimize the side effects, but infection, especially pneumonia, remains one of the main challenges in the early stage after kidney transplantation [2,3,4]. Over immunosuppression significantly impairs host immunity, leading to increased risk of infection. Currently, the routine surveillance in clinic is therapeutic drug monitoring [5, 6]. However, this strategy is quite restrictive and only provides indirect evidence of patient immune response. The high inter- and intra-patient variability and the deficiency of limited sampling strategies used in therapeutic drug monitoring increase the risk of graft failure [5, 7]. In contrast, the immune monitoring could provide direct information on patient response to immunosuppressive drugs or pathogens, thus contributing to better individualized therapy and long-term prognosis [8,9,10].
In recent years, a quantity of immune biomarkers have been found to diagnose or predict infection in solid organ transplant recipients, making them as parameters for immune monitoring. These include cell counts of lymphocytes, CD4+ T cells, CD8+ T cells and natural killer (NK) cells, CD4/CD8 ratio, molecule expression on specific cells such as human leukocyte antigen (HLA)-DR on monocytes, CD64 on neutrophils and programmed cell death protein 1 (PD-1) on lymphocytes, immunoglobulins, complements, soluble CD30 and immune cell response to stimuli [11,12,13]. However, in majority of these studies, parameters are usually analysed separately, or patients are simply classified by scoring according to the parameters. Single parameter without exact weight could not reflect the immune status accurately. Without comprehensive analysis by appropriate statistical methods, the efficacy of these parameters may not be ideal.
Compared with traditional methods, the machine learning techniques have advantages in big data processing. They have high power and accuracy, and can deal with numerous parameters simultaneously. Recently, these techniques have been used in therapeutic drug monitoring [14], and to predict the prognosis of chronic kidney disease [\(f\)(x) = SIGN (β0 + βmxm). When the result was 1, it was classified into the pneumonia group; when the result was -1, it was classified into the stable group. The function of LR model was \(f\)(x) = SIGMOID (β0 + βmxm). When the result was more than 0.5, it was classified into the pneumonia group; when the result was less than or equal to 0.5, it was classified into the stable group. As one of most common artificial neural networks widely used in machine learning tasks, the MLP model could be regarded as a logistic regression classifier with transformed features through several non-linear neural network layers. The RF model adopted ensemble learning technique by using multiple decision tree classifiers together to predict the result. A total of ten trees were developed. Each tree was presented with a different part of the dataset for training. The final prediction result was obtained through majority voting.
These machine learning models were also applied to predict the prognosis of pneumonia. Pneumonia patients were classified into mild group and severe group. Similar strategy was adopted as described above. After five rounds of training/validation rotation, the average sensitivity, specificity, PPV, NPV and AUC were calculated.
All the machine learning models were built using the programming language Python 3.6 and its machine learning library scikit-learn.
Statistical analysis
Continuous data were presented as the mean ± standard deviation (SD), and were compared using Student’s t-test, Welch’s t-test or the Mann–Whitney U test, where appropriate. Categorical data were compared using Pearson’s chi-squared (χ2) test or Fisher’s exact test, where appropriate. The performance of the models was assessed by calculating the area under the curve (AUC) of the receiver operating characteristic (ROC) curve. Statistical analysis was performed using SPSS version 22.0 (SPSS, Inc., Chicago, IL, USA). A P-value of < 0.05 was considered to be statistically significant.
Results
Basic characteristics
A total of 328 kidney transplant recipients underwent 955 tests of the immune monitoring panel from November 1st, 2017 to December 31st, 2019 in our center. A sizable part of the them were perioperative patients. Because the induction treatment had a significant impact on lymphocytes, only the patients more than or equal to three months post kidney transplantation were enrolled. The study flow was shown in Fig. 1.
46 eligible pneumonia patients underwent the immune monitoring test during the first week after admission to hospital. The average time from admission to test was 2.20 ± 2.08 days (2 patients received the test 7 days after admission and most cases received the first test within 3 days). Because they received the test when diagnosed with pneumonia, the average time from transplantation to receiving the test was the same as the time from transplantation to develo** pneumonia, namely 14.67 ± 15.24 months. Among them, 29 cases (63.0%) were between 3 and 12 months post transplantation, and 17 (37.0%) cases developed into severe pneumonia. As control, 100 eligible stable kidney transplant recipients with the data of 100 tests were randomly selected. The average time from transplantation to receiving the test was 10.33 ± 8.47 months.
The clinical characteristics of the pneumonia group and the stable group showed no significant difference in age, gender, donor source, time since transplant and CNI regimen. Obviously, the stable group had a better allograft function than the pneumonia group. Because 16 patients in the pneumonia group received transplants in other hospitals, the induction treatment was not available. As a result, the induction treatment showed significant difference in these two groups. The details were shown in Table 1.
Similarly, patients receiving the immune monitoring test from January 1st, 2020 to March 31st, 2020 were collected as external data for validation. 110 patients received 174 tests, but after exclusion, 10 pneumonia patients and 32 stable patients were enrolled. The characteristics of these patients were shown in Additional file 1: Table S2.
Immune status characterized by the panel
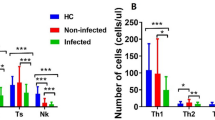
Compared with the stable group, the pneumonia group showed poor immune status, which was characterized by significantly lower cell counts of total T cells (CD3+ T cells), T cell subsets (CD4+ T cells and CD8+ T cells), B cells and NK cells (Table 2). Although the percentages of total T cells and NK cells showed statistical difference, they were not clinically significant (pneumonia vs stable, 76.79 ± 11.71vs 73.35 ± 10.28 for total T cells, P = 0.015; 12.78 ± 8.81 vs 17.11 ± 9.68 for NK cells, P = 0.003). The percentages of T cell subsets and B cells showed no significant difference. Notably, the CD4/CD8 ratio, which was reported as an immune biomarker, also showed no significant difference (pneumonia vs stable, 1.21 ± 0.61 vs 1.12 ± 0.59, P = 0.320).
The remaining two parameters also provided meaningful information. The expression of HLA-DR on monocytes was significantly lower in the pneumonia group (931.17 ± 671.15 vs 1392.53 ± 764.37, P < 0.001), while the expression of CD64 on neutrophils were much higher in the pneumonia group (589.20 ± 605.44 vs 101.11 ± 54.08, P < 0.001).
Machine learning models based on immune monitoring
To study whether the parameters of the immune monitoring panel were associated with pneumonia in kidney transplant recipients, univariate LR was performed to assess each parameter (Additional file 1: Table S3). Several parameters including monocyte HLA-DR, neutrophil CD64 and cell counts of T cells, B cells and NK cells showed significance, but the performance was not ideal (data not shown).
To improve the performance, machine learning models including SVM, LR, MLP and RF were developed as described in the methods. After five rounds of training/validation rotation, the average sensitivity, specificity, PPV, NPV and AUC of these modes were shown in Table 3. All the models had good results with AUC (Fig. 2), of which the SVM model had the highest AUC of 0.940. Notably, the SVM model also had good clinical practicality, with sensitivity of 81.7%, specificity of 92.0%, PPV of 83.6% and NPV of 91.3%. Monocyte HLA-DR (MFI), neutrophil CD64 (MFI), CD8+ T cells (cells/μl), NK cells (cell/μl) and TBNK (T cells, B cells and NK cells, cells/μl) were selected to build the SVM and LR models. The parameter coefficients were shown in Table 4. The MLP model, as one of the techniques of artificial neural network (ANN), calculated the probability of each category. The average AUC was 0.923, and the sensitivity, specificity, PPV and NPV were 71.8%, 92.0%, 82.7% and 87.9%, respectively. As an example, one tree of the RF model was shown in Fig. 3. A total of ten trees were developed. The final result was obtained through majority voting from the ten trees. The average AUC was 0.895, and the sensitivity, specificity, PPV and NPV were 73.6%, 95.0%, 88.0% and 89.2%, respectively.
The ROC curves and average AUC of the machine learning models. K-fold cross validation (k = 5) was used to estimate and compare the performance of different machine learning models. After five rounds of training/validation rotation, the average AUC was calculated. a The support vector machine (SVM) model. b The logistic regression (LR) model. c The multi-layer perceptron (MLP) model. d The random forest (RF) model. ROC curve, receiver operating characteristic curve. AUC area under the curve
Compared with mild pneumonia, severe pneumonia had a worse impact on allograft and patient survival. Among the 46 pneumonia patients, 17 cases progressed to severe pneumonia. Three patients died with functioning allografts, and one patient lost allograft. All of them were from the severe pneumonia group. Because all pneumonia patients received the immune monitoring tests early after admission (2.20 ± 2.08 days from admission to test), we also studied whether the result of the immune monitoring panel could predict the prognosis of pneumonia. The comparison between the two groups was shown in Table 5. Only the cell count of NK cells showed significance (135.60 ± 108.79 vs 59.28 ± 39.50, P = 0.027); the mild pneumonia group had higher monocyte HLA-DR, but not statistically significant (1068.59 ± 758.07 vs 696.76 ± 410.57, P = 0.127).
The machine learning models were used to predict the prognosis based on the immune monitoring panel, and the results were shown in Additional file 1: Table S4. The limited data had a negative impact on the effectiveness. The AUCs of SVM, LR, MLP and RF models were 0.600, 0.672, 0.716 and 0.760, respectively. Among them, the RF model had the best performance. A ten-tree RF model was developed, and one tree of the final algorithm was shown (Fig. 4a). Similarly, after five rounds of training/validation rotation, the average sensitivity, specificity, PPV and NPV of the RF model were 53.3%, 80.0%, 68.0% and 75.3%, respectively (ROC curve shown in Fig. 4b).
Validation by external data
To further confirm the association between the immune monitoring test and pneumonia in kidney transplant recipients, the machine learning models were validated by the external data, which contained 10 pneumonia patients and 32 stable patients. All the models had good performance, with accuracy over 80%. The SVM model had the highest AUC of 0.945, and the sensitivity, specificity, PPV, NPV and accuracy were 90.0%, 81.3%, 60.0%, 96.3% and 83.3%, respectively. The details of other models were shown in Additional file 1: Table S5.
Discussion
This study successfully provided a novel strategy to evaluate the significance of immune monitoring in kidney transplant recipients by machine learning models. Based on the results of immune monitoring panel, the SVM model best identified the kidney transplant recipients at risk of pneumonia, and the RF model best predicted the patients who would progress to severe pneumonia. All the models were validated by the external data, and showed good performance. The results of immune monitoring panel might contribute to better individualized therapy, including immunosuppressive drug adjustment and immunostimulant treatment.
Compared with traditional methods, the machine learning techniques could process multidimensional parameters simultaneously, and were not limited by data distribution [14]. Feature selection and parameter fitting were performed on training set, and evaluation of model performance was through validation set. Therefore, it had the ability of self-evolution by adjusting their structures when encountering errors [17]. The models could have better performance if more data obtained, making them promising in big data analysis. The immune system was exactly a very complicated network, and multiple parameters were needed to reflect the full picture of immune status. Therefore, the machine learning technique could be a powerful tool in analysis of immune monitoring.
The immune monitoring panel in this study consisted of the parameters that were relatively convenient and practical to obtain clinically. These included the percentages and absolute number of TBNK, HLA-DR on monocytes and CD64 on neutrophils. In our study, patients were not analysed by ATG and non-ATG groups due to the flaw of data from patients receiving transplantation out of our center. Because of the low ATG routine dose (1.00 mg/kg for 3 days) used in China, patients usually had lymphocyte reconstitution in three months (unpublished data). Therefore, we only enrolled patients more than three months post transplantation. Compared with the stable group, the pneumonia group had a much lower level of TBNK cell counts (including the respective subpopulations), lower expression of HLA-DR on monocytes but higher expression of CD64 on neutrophils. Fernández-Ruiz and colleagues reported that low TBNK cell counts in kidney transplant recipients predicted post-transplant opportunistic infection, and found that CD8+ T cells less than 100 cells/μl and CD4+ T cells less than 50 cells/μl at month 1 were the most valuable predictive parameters for non-ATG and ATG groups, respectively [2]. Luo and colleagues also reported lower cell counts and impaired function of CD4+ T cells, CD8+ T cells and NK cells in kidney transplant recipients with infection [19]. After analysis of machine learning models in our study, the cell counts of CD8+ T cells, NK cells and total TBNK were selected as parameters for SVM and LR model building, showing their importance in immune monitoring. The weight of these parameters was determined by the coefficients, which should be more accurate and reasonable than simple scoring [20, 21]. Notably, the percentages of TBNK subpopulations, including CD4/CD8 ratio, did not show significance. Similar result of CD4/CD8 ratio was observed in solid organ transplant patients, which showed poor diagnostic performance in infectious complications [22]. However, the inverted CD4/CD8 ratio (less than 1.0) was regarded as one of the parameters that defined immune risk phenotype in ESRD patients [21].
Monocyte HLA-DR and neutrophil CD64 were also important parameters in the models. Decreased expression of HLA-DR on monocytes was regarded as an unquestionable marker of monocyte anergy, which correlated with low cytokine release in response to bacterial challenges and reduced antigen presenting ability [11]. The clinical significance of monocyte HLA-DR has been verified in sepsis, and low monocyte HLA-DR expression was suggested as an indication for immunostimulant therapy [23]. In kidney transplant recipients, the expression of monocyte HLA-DR showed a significant decrease two weeks after transplantation compared with that before transplantation [24]. Monocyte HLA-DR also showed great difference between septic and non-septic groups after lung transplantation without modulating T cell reconstitution [25]. All these evidences proved that over immunosuppression not only impaired adaptive immunity, but also innate immunity.
The neutrophil CD64, on the contrary, remained low expression in the stable group. Once stimulated by inflammatory cytokines like interferon-γ (IFN-γ), a rapid and significant increase of expression of CD64 could be detected on neutrophils in 4 to 6 h, making it a sensitive indicator of systematic inflammation [26]. Importantly, immunosuppression treatment did not alter this characteristic. CD64 index even showed a better diagnostic performance of infectious complications than C reactive protein (CRP) or white blood cells in solid organ transplant patients [22]. Our study provided further evidence of clinical significance of neutrophil CD64 in kidney transplant recipients with pneumonia.
It must be noted that the algorithms derived from our study could not be utilized directly in other centers. It was because that the MFI of HLA-DR and CD64 in the models was relative value, which was determined not only by the expression intensity, but also by the setting of flow cytometer and the antibodies chosen. Only the MFI of fluorochrome PE, which had a fixed fluorochrome to antibody ratio, could be converted into absolute value of antibodies bound per cell (AB/C), making it possible for lab-to-lab standardization [11]. Another choice was using the internal reference microspheres, just like the commercial kit of Leuko64 (Trillium Diagnostics LCC, Meine, USA) to determine the CD64 index [22, 27]. Because this was not available in our center, MFI was directly used in the models. Nevertheless, the methodology to develop the models by machine learning could be adopted. Moreover, other valuable parameters, such as the concentration of immunoglobulins [28, 29] or complements [30, 31], could also be added to further improve the models [17].
Because this was a retrospective study, there were some limitations. The time point for the test of immune monitoring panel was not fixed, and the kinetic follow-up of immune monitoring was not available. The flaw of clinical data, including the definite etiological evidence, limited further stratification analysis. For machine learning techniques, the number of cases was relatively small. Further big data analysis or prospective cohort study was needed.
Conclusions
This study established machine learning models to confirm the association between immune monitoring and pneumonia in kidney transplant patients. The SVM model consisting of monocyte HLA-DR (MFI), neutrophil CD64 (MFI), CD8+ T cells (cells/μl), NK cells (cell/μl) and TBNK (cells/μl) best identified patients at risk of pneumonia. The RF model predicted the prognosis of pneumonia. In the era of big data, comprehensive analysis based on multi-dimensional parameters was an effective method to deeply understand the complexity of diseases. The machine learning technique provided a good choice, which was promising in data analysis and contributed to better individualized therapy.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AB/C:
-
Antibodies bound per cell
- ANN:
-
Artificial neural network
- APC:
-
Allophycocyanin
- APC-Cy7:
-
Allophycocyanin and cyanine dye 7
- ATG:
-
Anti-thymocyte globulin
- AUC:
-
Area under the curve
- BUN:
-
Blood urea nitrogen
- CNI:
-
Calcineurin inhibitor
- CRP:
-
C reactive protein
- DCD:
-
Donation after citizen’s death
- ESRD:
-
End-stage renal disease
- HLA:
-
Human leukocyte antigen
- IFN-γ:
-
Interferon-γ
- LR:
-
Logistic regression
- MFI:
-
Median fluorescence intensity
- MLP:
-
Multi-layer perceptron
- MMF:
-
Mycophenolate mofetil
- NK:
-
Natural killer
- NPV:
-
Negative predictive value
- PD-1:
-
Programmed cell death protein 1
- PE:
-
Phycoerythrin
- PerCP:
-
Peridinin-chlorophyll-protein
- PPV:
-
Positive predictive value
- RF:
-
Random forest
- ROC:
-
Receiver operating characteristic
- SD:
-
Standard deviation
- SVM:
-
Support vector machine
- TBNK:
-
T cells, B cells and NK cells
References
Rana A, Gruessner A, Agopian VG, Khalpey Z, Riaz IB, Kaplan B, Halazun KJ, Busuttil RW, Gruessner RW. Survival benefit of solid-organ transplant in the United States. JAMA Surg. 2015;150:252–9.
Fernandez-Ruiz M, Lopez-Medrano F, Allende LM, Andres A, Garcia-Reyne A, Lumbreras C, San-Juan R, Morales JM, Paz-Artal E, Aguado JM. Kinetics of peripheral blood lymphocyte subpopulations predicts the occurrence of opportunistic infection after kidney transplantation. Transpl Int. 2014;27:674–85.
Wiseman AC. Immunosuppressive Medications. Clin J Am Soc Nephrol. 2016;11:332–43.
Kotton CN, Kumar D, Caliendo AM, Huprikar S, Chou S, Danziger-Isakov L, Humar A. The Transplantation Society International CMVCG The third international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2018;102:900–31.
Shuker N, Shuker L, van Rosmalen J, Roodnat JI, Borra LC, Weimar W, Hesselink DA, van Gelder T. A high intrapatient variability in tacrolimus exposure is associated with poor long-term outcome of kidney transplantation. Transpl Int. 2016;29:1158–67.
Hu R, Barratt DT, Coller JK, Sallustio BC, Somogyi AA. Is there a temporal relationship between trough whole blood tacrolimus concentration and acute rejection in the first 14 days after kidney transplantation? Ther Drug Monit. 2019;41:528–32.
Hougardy JM, Maufort L, Cotton F, Coussement J, Mikhalski D, Wissing KM, Le Moine A, Broeders N, Abramowicz D. Therapeutic drug monitoring of enteric-coated mycophenolate sodium by limited sampling strategies is associated with a high rate of failure. Clin Kidney J. 2016;9:319–23.
Wekerle T, Segev D, Lechler R, Oberbauer R. Strategies for long-term preservation of kidney graft function. Lancet. 2017;389:2152–62.
Fernandez-Ruiz M, Kumar D, Humar A. Clinical immune-monitoring strategies for predicting infection risk in solid organ transplantation. Clin Transl Immunology. 2014;3:e12.
Khanna R. Immune monitoring of infectious complications in transplant patients: an important step towards improved clinical management. J Clin Microbiol. 2018;56:e01040.
Monneret G, Venet F. Sepsis-induced immune alterations monitoring by flow cytometry as a promising tool for individualized therapy. Cytometry B Clin Cytom. 2016;90:376–86.
Dendle C, Mulley WR, Holdsworth S. Can immune biomarkers predict infections in solid organ transplant recipients? A review of current evidence. Transplant Rev (Orlando). 2019;33:87–988.
Fernandez-Ruiz M, Lopez-Medrano F, Aguado JM. Predictive tools to determine risk of infection in kidney transplant recipients. Expert Rev Anti Infect Ther. 2020;18(5):423–41.
Zhang L, Zhang H, Ai H, Hu H, Li S, Zhao J, Liu H. Applications of machine learning methods in drug toxicity prediction. Curr Top Med Chem. 2018;18:987–97.
**ao J, Ding R, Xu X, Guan H, Feng X, Sun T, Zhu S, Ye Z. Comparison and development of machine learning tools in the prediction of chronic kidney disease progression. J Transl Med. 2019;17:119.
Tang J, Liu R, Zhang YL, Liu MZ, Hu YF, Shao MJ, Zhu LJ, **n HW, Feng GW, Shang WJ, et al. Application of machine-learning models to predict tacrolimus stable dose in renal transplant recipients. Sci Rep. 2017;7:42192.
Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–30.
Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM Jr, Musher DM, Niederman MS, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72.
Luo Y, **e Y, Zhang W, Lin Q, Tang G, Wu S, Huang M, Yin B, Huang J, Wei W, et al. Combination of lymphocyte number and function in evaluating host immunity. Aging (Albany NY). 2019;11:12685–707.
Blazik M, Hutchinson P, Jose MD, Polkinghorne KR, Holdsworth SR, Atkins RC, Chadban SJ. Leukocyte phenotype and function predicts infection risk in renal transplant recipients. Nephrol Dial Transplant. 2005;20:2226–30.
Crepin T, Gaiffe E, Courivaud C, Roubiou C, Laheurte C, Moulin B, Frimat L, Rieu P, Mousson C, Durrbach A, et al. Pre-transplant end-stage renal disease-related immune risk profile in kidney transplant recipients predicts post-transplant infections. Transpl Infect Dis. 2016;18:415–22.
Grey D, Sack U, Scholz M, Knaack H, Fricke S, Oppel C, Luderer D, Fangmann J, Emmrich F, Kamprad M. Increased CD64 expression on polymorphonuclear neutrophils indicates infectious complications following solid organ transplantation. Cytometry A. 2011;79:446–60.
Venet F, Lukaszewicz AC, Payen D, Hotchkiss R, Monneret G. Monitoring the immune response in sepsis: a rational approach to administration of immunoadjuvant therapies. Curr Opin Immunol. 2013;25:477–83.
Cho JH, Yoon YD, Jang HM, Kwon E, Jung HY, Choi JY, Park SH, Kim YL, Kim HK, Huh S, et al. Immunologic monitoring of T-lymphocyte subsets and Hla-Dr-positive monocytes in kidney transplant recipients: a prospective, observational cohort study. Medicine (Baltimore). 2015;94:e1902.
Alingrin J, Coiffard B, Textoris J, Nicolino-Brunet C, Gossez M, Jarrot PA, Dignat-George F, Monneret G, Thomas PA, Leone M, et al. Sepsis is associated with lack of monocyte HLA-DR expression recovery without modulating T-cell reconstitution after lung transplantation. Transpl Immunol. 2018;51:6–11.
Davis BH. Improved diagnostic approaches to infection/sepsis detection. Expert Rev Mol Diagn. 2005;5:193–207.
Muzlovic I, Ihan A, Stubljar D. CD64 index on neutrophils can diagnose sepsis and predict 30-day survival in subjects after ventilator-associated pneumonia. J Infect Dev Ctries. 2016;10:260–8.
Fernandez-Ruiz M, Lopez-Medrano F, Varela-Pena P, Lora-Pablos D, Garcia-Reyne A, Gonzalez E, Morales JM, San Juan R, Lumbreras C, Paz-Artal E, et al. Monitoring of immunoglobulin levels identifies kidney transplant recipients at high risk of infection. Am J Transplant. 2012;12:2763–73.
Augusto JF, Garnier AS, Demiselle J, Langs V, Picquet J, Legall R, Sargentini C, Culty T, Poli C, Ammi M, et al. Hypogammaglobulinemia and risk of severe infection in kidney transplant recipients. Transpl Infect Dis. 2016;18:741–51.
Fernandez-Ruiz M, Lopez-Medrano F, Varela-Pena P, Morales JM, Garcia-Reyne A, San Juan R, Lumbreras C, Lora-Pablos D, Polanco N, Andres A, et al. Hypocomplementemia in kidney transplant recipients: impact on the risk of infectious complications. Am J Transplant. 2013;13:685–94.
Sagedal S, Thiel S, Hansen TK, Mollnes TE, Rollag H, Hartmann A. Impact of the complement lectin pathway on cytomegalovirus disease early after kidney transplantation. Nephrol Dial Transplant. 2008;23:4054–60.
Acknowledgements
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (81771722 to Yingzi Ming).
Author information
Authors and Affiliations
Contributions
BP, KC and YM conceived and designed the study. HG collected the data. HT built the machine learning models. QZ and JL performed the immune monitoring tests. BP analyzed the data and wrote the manuscript. YM revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The research was conducted in accordance with the Declaration of Helsinki. The study was approved by the Institutional Review Board of Third **angya Hospital, Central South University (No. 2019-S448). The transplantation was in accordance with the 2007 Chinese “Regulation on Human Organ Transplantation”, and authorized by the DCD ethics committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1:
Additional tables.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Peng, B., Gong, H., Tian, H. et al. The study of the association between immune monitoring and pneumonia in kidney transplant recipients through machine learning models. J Transl Med 18, 370 (2020). https://doi.org/10.1186/s12967-020-02542-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-020-02542-2