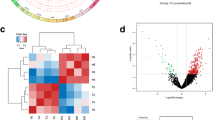
Abstract
Background
Circular RNAs (circRNAs) are a novel class of noncoding RNAs. Increasing evidence indicates that circRNAs play an important role in the occurrence and development of tumors. However, the role of circRNA hsa_circ_0044556 in the progression of colorectal cancer (CRC) remains unclear.
Methods
First, we searched for differentially expressed circRNAs using a circRNA microarray in paired CRC and adjacent normal tissues. The circRNA hsa_circ_0044556 was screened out from the existing CRC circRNA microarray in the Gene Expression Omnibus database and our microarray. The clinical significance of hsa_circ_0044556 expression level in CRC patients was then investigated. Finally, the functions of the targets of this circRNA were determined in CRC cell lines.
Results
Hsa_circ_0044556 was highly expressed in CRC patients and was positively correlated with tumor stage and lymph node metastasis. In CRC cell lines, the proliferation, migration, and invasion of cancer cells were inhibited by knocking down hsa_circ_0044556 expression.
Conclusion
Hsa_circ_0044556 promoted the progression of CRC. It is possible that hsa_circ_0044556 will become a novel biomarker or therapeutic target for CRC.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is a malignant cancer that seriously endangers the health of humans. Currently, the global incidence rate of CRC ranks third among cancer-related diseases with up to 1.2 million new cases each year, and more than 0.6 million deaths are expected each year [1]. From the perspective of the global incidence trend, the European and American regions are higher than the Asian and African regions, with the second place mortality rate (9.2%), and the develo** countries are higher than the developed countries [2]. In 2015, China's cancer statistics showed that the incidence and the mortality of CRC ranked fifth among cancer-related diseases in China. The incidence of CRC and the death toll are rising yearly while the population with this disease trends younger [3]. Early diagnosis, accurate prognosis, and recurrence monitoring play important roles in cancer diagnosis and treatment. The 5-year survival rate of patients with advanced CRC is only 12%, while the 5-year survival rate of patients at the early stage can reach more than 90% [4]. Therefore, early diagnosis can significantly improve the prognosis as well as the survival and quality of life of patients with CRC. On this basis, the in-depth study of the pathophysiology mechanisms underlying the occurrence and development of CRC will help us more comprehensively understand this cancer and open up new ideas and methods for the diagnosis and treatment of CRC. However, accurate and reliable prognosis and recurrence monitoring methods for CRC patients are still lacking in clinical practice, which makes it impossible to perform detailed posttreatment management and recurrence monitoring for patients [5, 6].
Circular RNAs (circRNAs) are a new class of noncoding RNAs (ncRNAs), which are single-stranded circular RNAs with no free 5′-end cap or 3′-end poly (A) tail. They are produced by alternative splicing of a specific pre-mRNA (pre-mRNA). Most circRNAs consist of exons and may also contain intergenic or noncoding regions [7,8,9,10]. CircRNAs did not attract much attention from researchers at first. For a long time, circRNAs were considered by-products of incorrect alternative splicing. Then in 2012, Salzman et al. discovered a large number of circRNAs using high-throughput sequencing technology [7]. Now it is recognized that circRNAs are not as rare as previously thought. In contrast, circRNAs are highly stable in cells with high expression, and sometimes their expression levels are even 10 times higher than those of their homologous messenger RNAs. It has been found that circRNAs can act as a miRNA sponge bound to RNA-binding protein to exert biological functions.
In recent years, circRNAs have been considered to play an important role in tumor progression [11]. CircRNAs are expected to become biomarkers for tumor diagnosis and prognosis due to their stable circular structure [12]. Xu et al. found that hsa_circ_0001649 was expressed at abnormally low levels in intrahepatic cholangiocarcinoma tissues and could promote cell proliferation and tumor metastasis [13]. Circ-ITCH can act as a sponge of miR-7 and miR-20a to inhibit the negative regulatory effect of the latter on the target gene ITCH, while ITCH has a tumor suppressor function by inhibition of the Wnt/β-catenin signaling pathway [14]. However, the role of circRNA hsa_circ_0044556 in CRC remains unclear and deserves further investigation.
Materials and methods
Tissue collection
In this study, tissue samples of 52 patients with CRC from the Third ** set predicted by databases of miRDB [Full size image
Bioinformatic analysis of the predicted genes in hsa_circ_0044556
Gene ontology (GO) analysis was performed on hsa_circ_0044556, and the functional roles of the top 10 most enriched target genes were investigated from the perspective of biological processes (Fig. 6a). The results showed that hsa_circ_0044556 had a strong correlation with the cell cycle, responses to steroid hormone stimulation, and angiogenesis. KEGG analysis of hsa_circ_0044556 indicated that its top nine enriched pathways included prostate cancer, the cancer pathway, and the ErbB signaling pathway (Fig. 6b). Seven target genes were enriched in the cancer pathway. These data suggested that hsa_circ_0044556 may play important roles in the malignant behavior of cancer by regulating the expression of target genes involved in these pathways.
Gene ontology (GO) enrichment analysis and KEGG pathway analysis for has_circ_0044556. a The top 10 significantly enriched target genes and their scores are listed as the x-axis and the y-axis, respectively. b The top 9 significantly enriched pathways and their scores were listed as the x-axis and the y-axis, respectively