Abstract
Background
Hepatocellular carcinoma (HCC) is a highly aggressive and heterogeneous disease. HCC cell lines established from different patients would be useful in elucidating the molecular pathogenesis. However, success of HCC primary culture establishment remains at low rate. We aim to establish and characterize HCC primary culture and the derived cell line.
Methods
Fresh tumor tissues were collected from 30 HCC patients. Culture conditions were optimized for the attachment and growth of the isolated hepatocytes. Granulin-epithelin precursor (GEP), a growth factor reported to associate with cancer stem cell properties, was examined by flow cytometry to elucidate its role on primary culture establishment. The primary cell line was characterized in detail.
Results
Cells isolated from 16 out of 30 HCC cases (53%) had viability more than 70% and were subject to subsequent in vitro culture. 7 out of 16 cases (44%) could give rise to cells that were able to attach and grow in culture. GEP expression levels significantly correlated with the viability of isolated hepatocytes and success rate of subsequent primary culture establishment. Cells from HCC patient 21 grew and expanded rapidly in vitro and was selected to be further characterized. The line, designated HCC21, derived from a Hong Kong Chinese female patient with HCC at Stage II. The cells exhibited typical epithelial morphology and expressed albumin, AFP and HBV antigens. The cell line was authenticated by short tandem repeat analysis. Comparative genome hybridization analysis revealed chromosomal loss at 1p35-p36, 1q44, 2q11.2-q24.3, 2q37, 4q12-q13.3, 4q21.21-q35.2, 8p12-p23, 15q11.2-q14, 15q24-q26, 16p12.1-p13.3, 16q, 17p, 22q and gain at 1q21-q43 in both HCC21 cells and the original clinical tumor specimen. Sequence analysis revealed p53 gene mutation. Subcutaneous injection of HCC21 cells into immunodeficient mice showed that the cells were able to form tumors at the primary injection sites and metastatic tumors in the peritoneal cavity.
Conclusions
The newly established cell line could serve as useful in vitro and in vivo models for studying primary HCC that possess metastasis ability.
Similar content being viewed by others
Background
Hepatocellular carcinoma (HCC) is one of the most common and aggressive human malignancies worldwide [1]. It is a highly heterogeneous disease in terms of its multiple molecular profiles and varied clinical outcomes [1]. The heterogeneous nature of HCC has impeded patient prognosis and treatment. Establishment of cell lines derived from different HCC patients is therefore useful for comprehensive study of pathogenesis of the disease. However, cell line establishment from fresh tumor tissues is technically difficult. Pre-operative procedures such as embolization and chemotherapy often induce extensive necrosis in tumor tissues and only small amount of viable tumor cells are available for subsequent culture. Besides, contamination and subsequent out-growth of infiltrating fibroblasts also made the cell line establishment more difficult. Therefore, the success rate of cell line establishment from fresh tumor tissues is extremely low [2],[3], and the situation is even worse for cell lines with metastatic potential [4].
Anoikis is another critical factor hindering cell line establishment from fresh tissues. Anoikis refers to the apoptosis in epithelial cells due to the loss of interaction with their extracellular matrix. During hepatocyte isolation, cells were detached from extracellular matrix. Such detachment was shown to induce apoptosis of the freshly isolated cells and greatly decreased the cell viability and success rate of subsequent primary culture [5]. Hepatocyte isolation involves the use of collagenase degrading collagen and other intercellular material that support the structure of liver [6], which could trigger anoikis in freshly isolated hepatocytes [7],[8]. Therefore, anoikis resistance is an important property that enables the freshly isolated cells to survive in the primary culture.
We previously demonstrated the granulin-epithelin precursor (GEP) was a hepatic oncofetal protein that defined a subpopulation of cancer stem cells (CSCs) in HCC [9]. It was proposed that CSCs possess certain properties that allow them to survive better than the other cell populations during primary culture [10]. However, the underlying mechanism was not well-understood. It was previously reported that GEP conferred anchorage-independence to cancer cells and protected them against anoikis [11],[12]. Therefore, GEP-expressing cells might have advantage to survive better during hepatocyte isolation through resistance to anoikis.
Cell lines are used extensively in biomedical research as in vitro models for the cell type being investigated. Validity of the data obtained from cell lines depends on their identities, and how closely they resemble the characteristics of corresponding original tumor. For cell line identity, it is revealed that the frequency of cell line misidentification is high. Recent studies showed that between 18 and 36% of cell lines were incorrectly designated [13],[14]. Accurate identification of cell lines is crucial during cell line development to avoid the risks of using misidentified cells. Short tandem repeat (STR) profiling has been recommended by the American Type Culture Collection Standards Development Organization (ATCC SDO) Workgroup ASN-0002 as the best method currently available for human cell line authentication [14],[15]. For resemblance with original tumors, cell lines have been criticized for their inherent instability upon long term culture. In addition, culture process may lead to selective growth of rapidly growing cells that have more molecular abnormalities. Therefore, it is suggested that routine cell line authentication back to the original tissues is needed to ensure that the cell lines are still representative of the tumors. However, donor tissues are not available for most of the established cell lines, making such authentication impossible. HCC cell line in the early passages therefore provides a better experimental model for studying hepatocarcinogenesis as it resembles more closely the original tumor.
In present study, we aimed to establish new HCC cell line from fresh tumor tissues and optimize the culture conditions to facilitate the cell line establishment. We attempted to investigate the role of GEP in the viability of the freshly isolated cells and the success rate of subsequent primary culture. Here, we showed that GEP level was positively correlated with the viability of freshly isolated hepatocytes and the success rate of subsequent primary culture. The culture conditions for the primary hepatocytes were optimized and a new cell line, designated HCC21, was established from the fresh tumor tissue of a Hong Kong female patient with early staged and moderately differentiated HCC. The line was authenticated, and its morphology, growth kinetics, migration ability, cytogenetic features, and in vivo tumorigenicity were characterized. This newly established cell line should serve as a useful model for studying the molecular pathogenesis of HCC.
Results
Primary culture establishment from fresh tumor tissues of 30 HCC patients
Fresh tumor tissues from 30 HCC patients were included in the primary culture establishment study. After enzymatic digestion by type IV collagenase, disaggregated cells were collected from the tumors. Cell viabilities were assessed and only cases with cell viability >70% were subject to subsequent in vitro culture. For cases #1 to #20, 10 out of 20 cases (50%) generated cells with >70% viability. Primary cells usually require extracellular matrix components such as collagen and fibronectin or biodegradable polymers such as gelatin to promote cell attachment. Of these, gelatin and collagen were reported to favour sustained viability and functions of hepatocytes [16]. Therefore, freshly isolated cells were resuspended in AMEM supplemented with 10% FBS and then seeded onto 6-well culture plates coated with either gelatin or collagen. Cells from 5 out of 10 cases (50%) attached to the culture plates coated with 0.1% gelatin; while only 2 out of 10 cases (20%) attached to those coated with 0.1% collagen. Cells growing on collagen-coated plate were sparse and die within 1 month, while those growing on gelatin-coated plate attached better and could establish cell-to-cell contacts (Figure 1A). Therefore, 0.1% gelatin was chosen to coat plate for primary cell culture of subsequent HCC cases.
Primary culture establishment from fresh tumor tissues of 30 HCC patients. Experimental protocol for establishing primary HCC culture from 30 patients was illustrated. Fresh tumor tissues were collected from HCC patients and underwent enzymatic digestion by type IV collagenase to release disaggregated cells. (A) For cases #1-20, cells were resuspended in AMEM supplemented with 10% FBS and then seeded onto 6-well culture plates coated with either 0.1% gelatin or 0.1% collagen. Phase contrast microscopy images of cultured cells from cases #14 and #17 were shown. (B) For cases #21-30, cells were resuspended in either AMEM supplemented with 10% FBS or HCM and then seeded onto 6-well culture plates coated with 0.1% gelatin. Phase contrast microscopy images of cultured cells from cases #21 and #29 were shown.
Fibroblast contamination and subsequent out-growth was frequently found in the process of cell line establishment. Therefore, we started to use defined media, hepatocyte culture medium (HCM), for cases #21 to #30. HCM provides a condition that permits preferential growth of hepatocytes, while that of contaminating fibroblasts is not favoured. For cases #21 to #30, 6 out of 10 cases (60%) generated cells with >70% viability, and the cells were resuspended in either AMEM supplemented with 10% FBS or HCM and then seeded onto 6-well culture plates coated with gelatin. Among the 6 cases, cells of #21 and #29 (2 out of 6 cases, 33%) could attach to culture plates coated with gelatin and grow in both 10% AMEM and HCM (Figure 1B). Fibroblast out-growth was observed in both cases cultured in AMEM with 10% FBS, while fibroblasts gradually decreased and disappeared in those cultured in HCM. Cells from #21 grew well and could propagate in subsequent passages, while those from #29 died gradually after one passage. Extensive characterization of cells from #21 was then performed.
Correlation of GEP expression with success rate of HCC primary culture
We previously demonstrated that GEP was a hepatic oncofetal protein that defined CSC population in HCC [9]. CSCs are proposed to possess certain properties that allow them to survive better in primary culture [10]. Further analysis with clinico-pathological features revealed that increased GEP level was significantly associated with poor differentiation (p = 0.044) (Additional file 1: Table S1). As tumors with poor differentiation were more aggressive and associated with more stem-cell features [17], the result implied that GEP might contribute to the aggressiveness and stemness of HCC, and GEP-expressing cells might survive better in primary culture.
We quantified the GEP levels of the original tumors by flow cytometry, and studied their relationship with the cell viability and success rate of cell line establishment. GEP level was shown to be significantly correlated with the viability of cells isolated from the fresh tumor tissues (n = 28, Spearman’s ρ correlation coefficient = 0.729, p = 0.000) (Figure 2A). Cells isolated from 7 cases showed viability >70% and could successfully grow in culture, and their GEP levels were significantly higher than those that failed to grow or the viability was <70% (p = 0.001, Figure 2B). The results suggested that GEP might be an important factor for facilitating cell line establishment from fresh HCC tumor tissues.
Correlation of GEP expression with success rate of HCC primary culture. (A) GEP levels significantly correlated with the viability of the cells freshly isolated from HCC tissues (n = 28, Spearman’s ρ correlation coefficient = 0.729, p = 0.000). GEP level was measured by flow cytometry and expressed as mean fluorescence intensity (MFI) after subtracting the non-specific background signal (isotype control). (B) GEP levels (MFI) of the cells with viability > 70% and could grew on culture plate successfully (viable and growth) were significantly higher than those failed or viability <70% (not viable or no growth) (n = 28, p = 0.001). (C) Hep3B cells were sorted for surface GEP expression by magnetic sorting. Unsorted control cells, sorted GEPhigh and GEPlow cells were then cultured in 10% FBS-supplemented AMEM in ultra-low attachment plate for 12 h. Cells were harvested and stained for annexin V-propidium iodide for apoptosis. Cells negative for both annexin V and propidium iodide were defined as viable cells, which were quantified by flow cytometric analysis. Viability of GEPlow cells was significantly lower than unsorted and sorted GEPhigh cells.
During hepatocyte isolation, cells were detached from extracellular matrix. Such detachment induced anoikis and greatly decreases the cell viability and success rate of subsequent primary culture [5]. GEP was shown to protect cancer cells against anoikis [11],[12], and might therefore facilitate primary culture establishment by conferring anoikis resistance to the HCC cells. To support this hypothesis, Hep3B cells were sorted based on GEP expression. Post-sorting analysis was performed and purity was shown to be >80% (Additional file 1: Figure S1). Unsorted control cells, sorted GEPhigh and GEPlow cells were then cultured in attachment free environment for 12 h. Viability of GEPlow cells was significantly lower than unsorted and sorted GEPhigh cells (p = 0.025 and p = 0.011, respectively), suggesting that GEP was crucial for protecting HCC cells from anoikis-induced apoptosis (Figure 2C) and might facilitate primary culture establishment.
Phenotypic characterization of HCC21 cells
The cells derived from HCC case #21, designated HCC21, reached 80% confluency at 10 days after initial culture, and sub-passage was performed successfully without significant cell death. The cells began to grow quickly at the 6th passage and were passaged for more than 50 generations thereafter. Flow cytometric analysis showed that albumin + cells of the original tumor #21 were 83.2%. Upon culture for about 10 days in hepatocyte culture medium, albumin + cells increased to 88.9%, which was further enriched to more than 98% after 6 passages (Figure 3A), confirming that HCC21 cells were hepatocytes. HCC21 cells grew as adherent monolayer with epithelial morphology (Figure 3B) and the cells maintained consistent morphology along passages. Growth curve of HCC21 cells is shown in Figure 3C. The population doubling time of HCC21 was approximately 95 h. Wound healing assay was performed to assess the migration ability of HCC21 cells. The cells started to migrate a day after the wound was made, and the wound healing was completed after 3 days (Figure 3D). The migration ability of HCC21 cells echoed the histology observation that venous infiltration (micrometastasis) was observed in the surgical specimen.
Phenotypic characterization of HCC21 cells. (A) Albumin levels of freshly isolated cells from patient’s tumor specimen, HCC21 cells at passage 1 and 6 were measured by flow cytometry. Percentages of albumin + cells were indicated in the histograms. (B) Phase-contrast microscopy images of cultured HCC21 cells at passages 6 and 30. (C) Growth curve of HCC21 cells at passage 10. (D) Wound healing assay showing the migration ability of HCC21 cells at passage 10.
STR profile analysis of HCC21 cells
The DNA samples of HCC21 early and late passages (11 and 50, respectively), original tumor and adjacent non-tumor liver tissue were subjected to DNA fingerprinting analysis. A total of 15 STR loci (CSF1P0, D2S1338, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D19S433, D21S11, FGA, TH01, TPOX, vWA) were co-amplified in each sample. AMLEO locus at the sex chromosomes was also examined. The data were analyzed and allele(s) of each locus were determined (Table 1). The STR profiles of HCC21 cells and the original tumor were identical, suggesting that they came from the same person. Besides, loss of heterozygosity (LOH) was observed in 2 loci, D16S539 and FGA in original tumor, and the aberrations were retained in HCC21 cells (Figure 4).
DNA fingerprinting of HCC21 cells. Comparison of genoty** results in adjacent non-tumor liver tissue and tumor specimen from patient, and HCC21 cells at passage 11 and 50. Patient’s tumor specimen and cultured HCC21 cells demonstrated loss of heterozygosity in (A) D16S539 and (B) FGA when compared to adjacent non-tumor liver tissue.
Array-based comparative genomic hybridization (aCGH) analysis
Changes in genomic copy number in HCC21 cells and original tumor were characterized by aCGH analysis. aCGH arrays were performed using DNA from original tumor and that from the HCC21 cells 6 months after its establishment. Chromosomal aberrations including chromosomal loss at 1p35-p36, 1q44, 2q11.2-q24.3, 2q37, 4q12-q13.3, 4q21.21-q35.2, 8p12-p23, 15q11.2-q14, 15q24-q26, 16p12.1-p13.3, 16q, 17p, 22q and gain at 1q21-q43 were observed in both the original tumor specimen and HCC21 cells (Figure 5). Noted the STR profiles showed LOH at FGA (at chromosome 4q28) and D16S539 (at chromosome 16q24.1), and that corroborated the aCGH data with chromosomal loss at 4q21.21-q35.2 and 16q in both the original clinical tumor specimen and the derived HCC21 cells.
CGH analysis of HCC21 cells and the original tumor. Vertical profiles of chromosomes 1, 2, 4, 8, 15, 16, 17 and 22 are shown with a log2 ratio, which represents the difference between tumor and normal DNA copy number. Ratio of 0 indicates that there is no change in copy number between tumor and normal cell DNA. Positive and negative log2 ratios indicate the gain and loss of tumor DNA copy number, respectively. The left panel of each plot represents the chromosome of the original clinical tumor specimen, while the right panel represents that of HCC21 cells at passage 10.
TP53 mutational analysis
p53 has been reported to be frequently mutated and overexpressed in HCCs [18]. Result from western blot showed that p53 protein was present in HCC21 cells. Human liver cancer cell lines HepG2 and PLC/PRF/5 were included as wild-type and mutant controls, respectively (Figure 6A). Subsequent analysis by DNA sequencing indicated a point mutation (Gly → Arg) (GGA → AGA) at codon 266 of exon 8 in both the HCC21 cells and tumor specimen (Figure 6B). PLC/PRF/5 was included as p53 mutant control (codon 249 of exon 7) and no mutation was found in exon 7 of HCC21 cells (data not shown).
TP53 mutational analysis in HCC21 cells. (A) Expression of p53 protein in HCC21 cells, HepG2 (wild-type p53) and PLC (p53 mutation at codon 249). (B) Sequencing analysis of TP53 gene revealed a point mutation (Gly→Arg) in codon 266 of exon 8 in HCC21 cells (passage 11) and patient’s tumor specimen, but not PLC.
Tumorigenicity and metastatic potential in immunodeficient mice
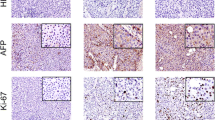
HCC21 cells were injected subcutaneously into 4 nude and 3 NOD/SCID mice. About 2 months after injection, visible tumors developed in all mice at the site of inoculation (Figure 7A), indicating that HCC21 cells were tumorigenic. We assessed the expression of HCC markers including AFP and HBV antigen [19] in the HCC21 cells and xenograft tumors. Western blot analysis showed that HCC21 cells and xenograft tumors in both nude and NOD/SCID mice retained the expression of HBV core antigen and AFP, which was consistent with original resected tumor (Figure 7B). Most importantly, secondary tumors were found in the peritoneal cavities of 2 NOD/SCID mice (Figure 8A). Histological sections of the xenografts showed neoplastic hepatocytes proliferation in broad trabeculae and acinar pattern, morphologically consistent with HCC. In addition, the metastatic (secondary) lesions were morphologically similar to the primary tumors (Figure 8B, upper panel). Besides, we have examined the expression of AFP in the primary and secondary xenograft tumors, in comparison to the patient’s tumor and adjacent non-tumor liver tissue specimen by immunohistochemical staining. Results showed that AFP protein was expressed at high levels in the tumor specimen, and also in the primary and secondary xenograft tumors. AFP protein could also be detected in adjacent non-tumor liver tissue specimen, but the signal was lower than the tumor counterpart (Figure 8B, middle panels). This further demonstrated that the tumor characteristics had been preserved from the original tumor specimen to the primary and metastasized tumor xenografts.
Tumorigenicity of HCC21 cells in immunocompromised mice. (A) Nude mice and NOD/SCID mice injected subcutaneously with 2x106 and 1x106 HCC21 cells, respectively, after 12 weeks. The middle panel shows the subcutaneous tumors derived from HCC21 cells 12 weeks after injection (scale bar = 10 mm). The lower panel shows the tumorgenicity of HCC21 cells. (B) Protein expression of HBV core antigen and AFP in patient’s tumor specimen, HCC21 xenograft tumors in NOD/SCID and nude mice, and cultured HCC21 cells at passage 12.
Metastatic potential of HCC21 cells in NOD/SCID mice. (A) Subcutaneous injection of HCC21 cells into NOD/SCID mice led to primary tumor development at the site of injection and secondary tumors in the peritoneal cavity (scale bar = 10 mm). (B) Histological analysis of patient’s tumor and adjacent non-tumor liver tissues, primary and secondary xenograft tumors established in NOD/SCID mice. Sections were stained with haematoxylin and eosin (H&E) (upper panel), AFP (middle panel) and rabbit Ig control (lower panel). (C) Undetectable protein expression of E-cadherin in HCC21 (passage 12) while HepG2 served as positive control.
The aCGH analysis revealed loss of 16q in HCC21 cells, which region contained E-cadherin gene (at chromosome 16q22.1). E-cadherin is an adhesion molecule crucial for inhibiting metastasis by connecting homophilic cells [20]-[22]. Loss of E-cadherin was reported to associate with high incidence of lymph node metastasis in various cancers, including HCC [23]-[25]. Therefore, we examined the expression of E-cadherin in HCC21 cells by western blot. HepG2, HCC cell line without metastatic potential, was included as positive control for the presence of E-cadherin. Result showed that E-cadherin protein was absent in HCC21 cells (Figure 8C), thereby providing further evidence for the metastatic potential of the cells.
Discussion
HCC is a highly heterogeneous in terms of molecular profiles and clinical outcomes [1]. Establishment of primary HCC culture derived from different patients is therefore useful for comprehensive study of the etiology and molecular pathogenesis of disease. However, establishment of human cell lines from fresh patients’ tumors is technically difficult, partly due to the low viability of the freshly isolated HCC cells. Here, we showed for the first time that GEP protein levels of HCC cells were significantly correlated to the cell viability and also the success rate of subsequent primary culture. GEP-expressing cells were able to survive better during hepatocyte isolation, which was potentially due to their resistance to anoikis. The findings therefore suggested that GEP might be an important factor for facilitating cell line establishment from fresh HCC tumor tissues.
Recent reports on GEP binding partners might provide some hints on the underlying mechanism for GEP-mediated anoikis resistance. GEP was demonstrated as a ligand of tumor necrosis factor receptors (TNFRs) in human chondrocytes and disturbed TNFα signaling [26]. Notably, TNFR1 has been shown to be crucial on anoikis execution in cancer progression [27]. In addition, our group have recently demonstrated that GEP interacted with heparan sulfate on HCC cell surface [28]. Heparan sulfate was essential for anoikis resistance and survival in mouse dermal fibroblasts [29] and in rat embryo fibroblasts [30]. We have further shown that tropomyosin 3 was the intracellular binding partner of GEP in liver cancer cells [31]. Independently, tropomyosin-1 was reported to regulate anoikis in breast cancer cells [32]. Therefore, further investigations are warranted to investigate the roles of TNFR, heparan sulfate and tropomyosins on GEP-mediated anoikis resistance in human cancer cells.
Cell line authentication is crucial during cell line development to avoid the risks of using misidentified cells. Recent studies have shown that more than 360 cell lines were cross-contaminated or misidentified without authenticated stock, and the validity of the studies using these cell lines was in doubt [33]. A cell line is considered misidentified when its DNA profile is not consistent with the individual from whom it was derived from. Here, we showed that the STR profiles of HCC21 cells and original resected tumor were identical, confirming that HCC21 cells and the tumor came from the same person.
Chromosomal alterations in HCC are complex [34], and this has impeded the elucidation of molecular mechanisms underlying hepatocarcinogenesis. Chromosomal aberrations in HCC21 cells were examined using CGH. The results showed that chromosomal aberrations were observed in both the primary tumor and HCC21 derived cells including loss at 1p35-p36, 1q44, 2q11.2-q24.3, 2q37, 4q12-q13.3, 4q21.21-q35.2, 8p12-p23, 15q11.2-q14, 15q24-q26, 16p12.1-p13.3, 16q, 17p, 22q and gain at 1q21-q43. Of these, gain at 1q was previously reported to be associated with HCC development [35]-[37]. Loss of 4q11-23 was previously reported to be observed in tumor larger than 3 cm. This is consistent with the tumor size of patient (8.5 cm). Chromosome 4q abnormality was significantly associated with HCC [38],[39]. LOH at 4q was reported to strongly correlate with AFP elevation in HCC [Short tandem repeat (STR) analysis DNA samples of the HCC21 cells at passage 11 and 50, original tumor and the adjacent non-tumor liver tissue from the patient were subjected to DNA fingerprinting analysis using the AmpF/STR Identifiler Plus PCR Amplification Kit (Invitrogen). A total of 15 STR loci (CSF1P0, D2S1338, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D19S433, D21S11, FGA, TH01, TPOX, vWA) were co-amplified in each sample and detected on an ABI 3130 Genetic Analyzer. AMLEO locus at the sex chromosomes was also examined. The data were analyzed and allele(s) of each locus were determined by GeneScan and GeneMapperTM ID Software (Invitrogen). Genomic DNA was extracted from HCC21 cells and the original tumor. The Agilent human 44 K CGH microarray (Agilent Technologies, Santa Clara, CA) with an average probe spatial resolution of ~ 35 kb was utilized. Genomic DNA at 1 μg was labelled by the Agilent Genomic DNA Labeling Kit PLUS (Agilent Technologies). The hybridized array was scanned by an Agilent Microarray Scanner and data were analysed by Agilent Feature Extraction 9.1 followed by computation and normalization using CGH Analytics v3.4. Putative chromosome copy number loss was defined by intervals of two or more adjacent probes with log2 ratios suggestive of a deletion when compared with the log2 ratios of adjoining probes. The Quality-Weighted Interval Score algorithm (ADM2) with a value of 6.0 was used to compute and assist the identification of aberrations for a given sample. Total protein was extracted with cell lysis buffer (Cell Signaling Technology, Boston, MA) in the presence of complete protease inhibitor cocktail (Roche, Mannheim, Germany) and separated in 8-10% SDS PAGE gel. Proteins were then electro-transferred onto polyvinylidene difluoride membranes, subsequently incubated with primary anti-human antibodies, detection by horseradish peroxidase-labeled secondary antibodies, and visualized with Enhanced Chemiluminescence Western Blotting Detection Kit (Amersham Biosciencse, Piscataway, NJ). Direct DNA sequencing was performed for exons 4-9 of p53, in which >80% of all mutations were observed [54],[55]. Primer sets and reaction conditions were adopted from Lehman et al[56]. DNA was amplified by polymerase chain reaction and direct DNA sequencing was performed with the BigDye Sequencing kit (Invitrogen). Electrophoresis and sequence analysis were performed using the ABI PRISM 3100 (Invitrogen). The study protocol was approved by and performed in accordance with the Committee of the Use of Live Animals in Teaching and Research at the University of Hong Kong. Cells from passage 8 were harvested, washed, and resuspended in hepatocyte culture medium (HCM). 1x106 cells were injected subcutaneously into the right flank of each athymic nude or NOD/SCID mouse (4 weeks old). The mice were examined every week for the development of tumors and tumor-bearing mice were sacrificed when tumor burden exceed 10% of the normal body weight or the body weight loss exceed 20%. Immunohistochemistry was performed with the Dako Envision Plus System (Dako, Carpinteria, CA) following the manufacturer’s instruction with modifications. Briefly, antigen retrieval was performed by microwave with sections immersed in citrate buffer. Followed by endogenous peroxidase blocking, tissues were stained with rabbit anti-human AFP antibody (Dako) or rabbit Ig (R&D Systems, Minneapolis, MN). The signal was detected by horseradish peroxidase-conjugated secondary antibody and color was developed with diaminobenzidine as the chromogen. The tissue sections were then counterstained with hematoxylin. All data were expressed as mean values + standard deviation (SD) from at least three independent experiments. Differences between groups were assessed by the Student’s t test. A probability (p) < 0.05 was considered significantly different. All analyzes were performed using the statistical software GraphPad Prism for Windows, Version 3.00 (GraphPad Software, CA).Array-based comparative genomic hybridization (aCGH)
Western blot analysis
TP53 mutational analysis
In vivo tumorigenicity in immunodeficient mice
Immunohistochemical staining
Statistical analyzes
Additional file
Abbreviations
- aCGH:
-
Array-based comparative genomic hybridization
- CSC:
-
Cancer stem cell
- GEP:
-
Granulin-epithelin precursor
- HCC:
-
Hepatocellular carcinoma
- STR:
-
Short tandem repeat (STR)
References
El-Serag HB: Hepatocellular carcinoma. N Engl J Med. 2011, 365 (12): 1118-1127. 10.1056/NEJMra1001683.
Hambly RJ, Double JA, Thompson MJ, Bibby MC: Establishment and characterisation of new cell lines from human breast tumours initially established as tumour xenografts in NMRI nude mice. Breast Cancer Res Treat. 1997, 43 (3): 247-258. 10.1023/A:1005756632293.
Park JG, Lee JH, Kang MS, Park KJ, Jeon YM, Lee HJ, Kwon HS, Park HS, Yeo KS, Lee KU, Kim ST, Chung JK, Hwang YJ, Lee HS, Kim CY, Lee YI, Chen TR, Hay RJ, Song SY, Kim WH, Kim CW, Kim YI: Characterization of cell lines established from human hepatocellular carcinoma. Int J Cancer. 1995, 62 (3): 276-282. 10.1002/ijc.2910620308.
Dangles-Marie V, Pocard M, Richon S, Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N, Validire P, Dutrillaux B, Praz F, Bellet D, Poupon MF: Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007, 67 (1): 398-407. 10.1158/0008-5472.CAN-06-0594.
Grossmann J, Walther K, Artinger M, Kiessling S, Scholmerich J: Apoptotic signaling during initiation of detachment-induced apoptosis ("anoikis") of primary human intestinal epithelial cells. Cell Growth Differ. 2001, 12 (3): 147-155.
Seglen PO: Preparation of isolated rat liver cells. Methods Cell Biol. 1976, 13: 29-83. 10.1016/S0091-679X(08)61797-5.
Hansen LK, Mooney DJ, Vacanti JP, Ingber DE: Integrin binding and cell spreading on extracellular matrix act at different points in the cell cycle to promote hepatocyte growth. Mol Biol Cell. 1994, 5 (9): 967-975. 10.1091/mbc.5.9.967.
Smets FN, Chen Y, Wang LJ, Soriano HE: Loss of cell anchorage triggers apoptosis (anoikis) in primary mouse hepatocytes. Mol Genet Metab. 2002, 75 (4): 344-352. 10.1016/S1096-7192(02)00004-5.
Cheung PF, Cheng CK, Wong NC, Ho JC, Yip CW, Lui VC, Cheung AN, Fan ST, Cheung ST: Granulin-epithelin precursor is an oncofetal protein defining hepatic cancer stem cells. PLoS One. 2011, 6 (12): e28246-10.1371/journal.pone.0028246.
van Staveren WC, Solis DY, Hebrant A, Detours V, Dumont JE, Maenhaut C: Human cancer cell lines: experimental models for cancer cells in situ? For cancer stem cells?. Biochim Biophys Acta. 2009, 1795 (2): 92-103.
He Z, Ismail A, Kriazhev L, Sadvakassova G, Bateman A: Progranulin (PC-cell-derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res. 2002, 62 (19): 5590-5596.
Ong CH, Bateman A: Progranulin (granulin-epithelin precursor, PC-cell derived growth factor, acrogranin) in proliferation and tumorigenesis. Histol Histopathol. 2003, 18 (4): 1275-1288.
Lacroix M: Persistent use of "false" cell lines. Int J Cancer. 2008, 122 (1): 1-4. 10.1002/ijc.23233.
Workgroup ATCCSDO: Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010, 10 (6): 441-448. 10.1038/nrc2852.
Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, Furtado M, Kline MC, Kohara A, Los GV, MacLeod RA, Masters JR, Nardone M, Nardone RM, Nims RW, Price PJ, Reid YA, Shewale J, Sykes G, Steuer AF, Storts DR, Thomson J, Taraporewala Z, Alston-Roberts C, Kerrigan L: Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues. In Vitro Cell Dev Biol Anim. 2010, 46 (9): 727-732. 10.1007/s11626-010-9333-z.
Li K, Qu X, Wang Y, Tang Y, Qin D, Feng M: Improved performance of primary rat hepatocytes on blended natural polymers. J Biomed Mater Res A. 2005, 75 (2): 268-274. 10.1002/jbm.a.30415.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA: An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008, 40 (5): 499-507. 10.1038/ng.127.
Liu J, Ma Q, Zhang M, Wang X, Zhang D, Li W, Wang F, Wu E: Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis. Eur J Cancer. 2012, 48 (15): 2328-2338. 10.1016/j.ejca.2012.03.001.
Kim do Y, Han KH: Epidemiology and surveillance of hepatocellular carcinoma. Liver Cancer. 2012, 1 (1): 2-14. 10.1159/000339016.
Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, Peterse JL, Cardiff RD, Berns A, Jonkers J: Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006, 10 (5): 437-449. 10.1016/j.ccr.2006.09.013.
Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G: A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998, 392 (6672): 190-193. 10.1038/32433.
Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeier W: E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991, 113 (1): 173-185. 10.1083/jcb.113.1.173.
Winter JM, Ting AH, Vilardell F, Gallmeier E, Baylin SB, Hruban RH, Kern SE, Iacobuzio-Donahue CA: Absence of E-cadherin expression distinguishes noncohesive from cohesive pancreatic cancer. Clin Cancer Res. 2008, 14 (2): 412-418. 10.1158/1078-0432.CCR-07-0487.
Joo YE, Rew JS, Park CS, Kim SJ: Expression of E-cadherin, alpha- and beta-catenins in patients with pancreatic adenocarcinoma. Pancreatology. 2002, 2 (2): 129-137. 10.1159/000055903.
Hashiguchi M, Ueno S, Sakoda M, Iino S, Hiwatashi K, Minami K, Ando K, Mataki Y, Maemura K, Shinchi H, Ishigami S, Natsugoe S: Clinical implication of ZEB-1 and E-cadherin expression in hepatocellular carcinoma (HCC). BMC Cancer. 2013, 13 (1): 572-10.1186/1471-2407-13-572.
Tang W, Lu Y, Tian QY, Zhang Y, Guo FJ, Liu GY, Syed NM, Lai Y, Lin EA, Kong L, Su J, Yin F, Ding AH, Zanin-Zhorov A, Dustin ML, Tao J, Craft J, Yin Z, Feng JQ, Abramson SB, Yu XP, Liu CJ: The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011, 332 (6028): 478-484. 10.1126/science.1199214.
Paoli P, Giannoni E, Chiarugi P: Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013, 1833 (12): 3481-3498. 10.1016/j.bbamcr.2013.06.026.
Yip CW, Cheung PF, Leung IC, Wong NC, Cheng CK, Fan ST, Cheung ST: Granulin-epithelin precursor interacts with heparan sulfate on liver cancer cells. Carcinogenesis. 2014, pii: bgu164-Epub ahead of print
Verderio EA, Telci D, Okoye A, Melino G, Griffin M: A novel RGD-independent cell adhesion pathway mediated by fibronectin-bound tissue transglutaminase rescues cells from anoikis. J Biol Chem. 2003, 278 (43): 42604-42614. 10.1074/jbc.M303303200.
Jeong J, Han I, Lim Y, Kim J, Park I, Woods A, Couchman JR, Oh ES: Rat embryo fibroblasts require both the cell-binding and the heparin-binding domains of fibronectin for survival. Biochem J. 2001, 356 (Pt 2): 531-537. 10.1042/0264-6021:3560531.
Lam CY, Yip CW, Poon TC, Cheng CK, Ng EW, Wong NC, Cheung PF, Lai PB, Ng IO, Fan ST, Cheung ST: Identification and characterization of tropomyosin 3 associated with granulin-epithelin precursor in human hepatocellular carcinoma. PLoS One. 2012, 7 (7): e40324-10.1371/journal.pone.0040324.
Bharadwaj S, Thanawala R, Bon G, Falcioni R, Prasad GL: Resensitization of breast cancer cells to anoikis by tropomyosin-1: role of Rho kinase-dependent cytoskeleton and adhesion. Oncogene. 2005, 24 (56): 8291-8303. 10.1038/sj.onc.1208993.
Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI: Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010, 127 (1): 1-8. 10.1002/ijc.25242.
Lau SH, Guan XY: Cytogenetic and molecular genetic alterations in hepatocellular carcinoma. Acta Pharmacol Sin. 2005, 26 (6): 659-665. 10.1111/j.1745-7254.2005.00126.x.
Wong N, Lam WC, Lai PB, Pang E, Lau WY, Johnson PJ: Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. Am J Pathol. 2001, 159 (2): 465-471. 10.1016/S0002-9440(10)61718-X.
Chen YJ, Yeh SH, Chen JT, Wu CC, Hsu MT, Tsai SF, Chen PJ, Lin CH: Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology. 2000, 119 (2): 431-440. 10.1053/gast.2000.9373.
Qin LX, Tang ZY, Sham JS, Ma ZC, Ye SL, Zhou XD, Wu ZQ, Trent JM, Guan XY: The association of chromosome 8p deletion and tumor metastasis in human hepatocellular carcinoma. Cancer Res. 1999, 59 (22): 5662-5665.
Piao Z, Park C, Park JH, Kim H: Deletion map** of chromosome 4q in hepatocellular carcinoma. Int J Cancer. 1998, 79 (4): 356-360. 10.1002/(SICI)1097-0215(19980821)79:4<356::AID-IJC8>3.0.CO;2-U.
Yeh SH, Chen PJ, Lai MY, Chen DS: Allelic loss on chromosomes 4q and 16q in hepatocellular carcinoma: association with elevated alpha-fetoprotein production. Gastroenterology. 1996, 110 (1): 184-192. 10.1053/gast.1996.v110.pm8536855.
Zhang H, Ma H, Wang Q, Chen M, Weng D, Wang H, Zhou J, Li Y, Sun J, Chen Y, Liang X, Zhao J, Pan K, Wang H, **a J: Analysis of loss of heterozygosity on chromosome 4q in hepatocellular carcinoma using high-throughput SNP array. Oncol Rep. 2010, 23 (2): 445-455.
Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K: Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999, 29 (6): 1858-1862. 10.1002/hep.510290636.
Guan XY, Sham JS, Tai LS, Fang Y, Li H, Liang Q: Evidence for another tumor suppressor gene at 17p13.3 distal to TP53 in hepatocellular carcinoma. Cancer Genet Cytogenet. 2003, 140 (1): 45-48. 10.1016/S0165-4608(02)00654-4.
Vogelstein B, Kinzler KW: p53 function and dysfunction. Cell. 1992, 70 (4): 523-526. 10.1016/0092-8674(92)90421-8.
Ozaki T, Nakagawara A: p53: the attractive tumor suppressor in the cancer research field. J Biomed Biotechnol. 2010, 2011: 603925-
Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M: Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007, 28 (6): 622-629. 10.1002/humu.20495.
Wang L, Yao M, Dong Z, Zhang Y, Yao D: Circulating specific biomarkers in diagnosis of hepatocellular carcinoma and its metastasis monitoring. Tumour Biol. 2014, 35 (1): 9-20. 10.1007/s13277-013-1141-0.
Tian J, Tang ZY, Ye SL, Liu YK, Lin ZY, Chen J, Xue Q: New human hepatocellular carcinoma (HCC) cell line with highly metastatic potential (MHCC97) and its expressions of the factors associated with metastasis. Br J Cancer. 1999, 81 (5): 814-821. 10.1038/sj.bjc.6690769.
Alexander JJ, Bey EM, Geddes EW, Lecatsas G: Establishment of a continuously growing cell line from primary carcinoma of the liver. S Afr Med J. 1976, 50 (54): 2124-2128.
Voskoglou-Nomikos T, Pater JL, Seymour L: Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003, 9 (11): 4227-4239.
Kalluri R, Weinberg RA: The basics of epithelial-mesenchymal transition. J Clin Invest. 2009, 119 (6): 1420-1428. 10.1172/JCI39104.
Kang Y, Massague J: Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004, 118 (3): 277-279. 10.1016/j.cell.2004.07.011.
Kessenbrock K, Plaks V, Werb Z: Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010, 141 (1): 52-67. 10.1016/j.cell.2010.03.015.
Ho JC, Ip YC, Cheung ST, Lee YT, Chan KF, Wong SY, Fan ST: Granulin-epithelin precursor as a therapeutic target for hepatocellular carcinoma. Hepatology. 2008, 47 (5): 1524-1532. 10.1002/hep.22191.
Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC: TP53 and liver carcinogenesis. Hum Mutat. 2003, 21 (3): 201-216. 10.1002/humu.10176.
Hollstein M, Sidransky D, Vogelstein B, Harris CC: p53 mutations in human cancers. Science. 1991, 253 (5015): 49-53. 10.1126/science.1905840.
Lehman TA, Bennett WP, Metcalf RA, Welsh JA, Ecker J, Modali RV, Ullrich S, Romano JW, Appella E, Testa JR, Gerwin BI, Haris CC: p53 mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer Res. 1991, 51 (15): 4090-4096.
Acknowledgements
This study was supported in part by Sun C.Y. Research Foundation for Hepatobiliary and Pancreatic Surgery, Hong Kong Research Grants Council (764111, 764112 and T12-401/13-R) and Health and Medical Research Fund (01121566).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
S.T. Cheung has received Pfizer collaborative research grants. The other authors disclose no conflicts. The sponsors had no role in study design as well as in data collection, analysis, and interpretation.
Authors’ contributions
PFYC carried out experiments, analyzed data and wrote the manuscript. STC participated in study design, analyzed data, obtained funding and revised the manuscript. CWY, and LWCN carried out the experimental work and analyzed the data. TTC, RTPP, and STF provided clinical specimens and information. KWL and CC performed and analyzed STR data. NW and KWC performed and analyzed aCGH data. KFC performed histological analysis. All authors read, commented and approved the final manuscript.
Electronic supplementary material
12935_2014_103_MOESM1_ESM.doc
Additional file 1: Figure S1. Post-sorting analysis on GEP expression in unsorted Hep3B and freshly isolated GEPhigh, and GEPlow subpopulations. Table S1. Clinicopathological features of HCC in relation to GEP expression. (DOC 92 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Cheung, P.FY., Yip, C.W., Ng, L.WC. et al. Establishment and characterization of a novel primary hepatocellular carcinoma cell line with metastatic ability in vivo. Cancer Cell Int 14, 103 (2014). https://doi.org/10.1186/s12935-014-0103-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-014-0103-y