Abstract
Background
Cryptosporidium spp. and Giardia duodenalis are important gastrointestinal protists in humans and animals worldwide. In China, bovine cryptosporidiosis and giardiasis are of increasing concern because cattle are important reservoirs of these parasites, which have become potential threats to public health and to large numbers of cattle in recent years.
Results
A total of 1366 fecal samples from the Ningxia Autonomous Region were examined. The overall infection rates for Cryptosporidium spp. and G. duodenalis were 1.61% and 2.12%, respectively. Cryptosporidium was only detected in preweaned calves and adults older than 2 years, whereas G. duodenalis was only detected in calves aged less than 11 months. Cryptosporidium spp. were characterized with a PCR–restriction fragment length polymorphism analysis and DNA sequence analysis of the small subunit rRNA gene. Three Cryptosporidium species were identified: C. parvum (n = 15) and C. bovis (n = 4) in preweaned calves, and C. andersoni (n = 4) in adults aged over 2 years. A DNA sequence analysis of the gp60 gene suggested that the 15 C. parvum isolates all belonged to subtype IIdA15G1. Twenty-nine G. duodenalis isolates were analyzed by DNA sequencing of the triosephosphate isomerase (tpi) and glutamate dehydrogenase (gdh) genes. Two G. duodenalis assemblages were identified, assemblages E (n = 15) and B (n = 4, one subtype B1 and three subtype B2) in preweaned calves, and assemblage E (n = 10) in 3–11-month-old calves.
Conclusions
The predominance of C. parvum detected in preweaned calves and the first identified subtype IIdA15G1 in dairy cattle, and the dominant G. duodenalis assemblage E in this study differed considerably from those found in Henan, Heilongjiang, and Shannxi Provinces. Our findings further confirm the dominance of C. parvum IId subtypes in China.
Similar content being viewed by others
Background
Cryptosporidium and Giardia are important gastrointestinal protists with a wide spectrum of hosts, including humans, livestock, companion animals, and wildlife. Infection is acquired via the fecal–oral route following the ingestion of infective oocysts or cysts, by either direct contact or the ingestion of contaminated food or water [12]. Thus, the distributions of Cryptosporidium spp. and the C. parvum subtype (all belonging to IIdA19G1) in dairy cattle differ considerably from those in other countries [13]. The objective of this study was to identify the species of Cryptosporidium and Giardia present in dairy cattle in the Ningxia Autonomous Region, northwest China.
Methods
Ethics statement
This study was conducted in accordance with the Chinese Laboratory Animal Administration Act of 1988. The research protocol was reviewed and approved by the Research Ethics Committee of the Henan Agricultural University. Permission was obtained from farm owners before collection of fecal samples.
Sample collection and examination
A fresh fecal sample was collected from each animal using a sterile disposal latex glove immediately after its defecation onto the ground, and was then placed individually into a disposable plastic bag. In total, 1366 fecal samples were collected between December 2011 and October 2012 from dairy cattle on 20 scale farms in the Ningxia Autonomous Region, northwestern China (Table 1). The Cryptosporidium oocysts in the fecal materials were concentrated with Sheather’s sugar flotation technique, with a further formalin–ethyl acetate sedimentation step included for the preweaned calf samples [9]. Giardia cysts were detected with Lugol’s iodine staining. Cryptosporidium- or Giardia-positive samples were stored in 2.5% potassium dichromate at 4°C before DNA extraction.
DNA extraction
The Cryptosporidium or Giardia-positive fecal specimens were washed three times with distilled water, and the genomic DNA was extracted from the fecal pellets with the E.Z.N.A.® Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA), according to the manufacturer’s recommendations.
Cryptosporidium/Giardiagenoty** and subty**
The Cryptosporidium species were identified with PCR–restriction fragment length polymorphism (RFLP) analysis and DNA sequence analysis of the small subunit (SSU) rRNA gene [17], and the genotype/subtype identities of the G. duodenalis samples were established by direct comparison of these sequences with reference sequences downloaded from the GenBank database.
DNA sequence analysis
PCR products were sequenced on an ABI Prism™ 3730 XL DNA Analyzer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster, CA, USA). Sequence accuracy was confirmed with bi-directional sequencing and by sequencing a new PCR product if necessary. The sequences were aligned with the ClustalX 1.83 program. Representative nucleotide sequences have been deposited in GenBank under accession numbers KM067089–KM067096.
Statistical analysis
The χ2 test was used to compare the Cryptosporidium infection rates. Differences were considered significant at P <0.05.
Results
Prevalence of Cryptosporidium and Giardia
Microscopic analysis of 1366 fecal samples showed the presence of Cryptosporidium oocysts in 23 samples (1.68%) from nine farms, and the highest infection rate was 9.38% on farm 2 (Table 1). The infection rates of Cryptosporidium spp. were 10.22%, 0%, 0%, and 0.69% in preweaned, 3–11-month-old, 1–2-year-old, and > 2-year-old cattle, respectively (χ2 = 95.52, P <0.01).
Similarly, 29 samples from 12 farms were positive for Giardia, with an average infection rate of 2.12% (Table 1). The highest infection rate for Giardia was 7.94% on farm 5. The infection rates for Giardia were 10.22%, 2.53%, 0%, and 0% in preweaned, 3–11-month-old, 1–2-year-old, and > 2-year-old cattle, respectively (χ2 = 75.93, P <0.01).
Distribution of Cryptosporidium species/subtypes and G. duodenalisassemblages
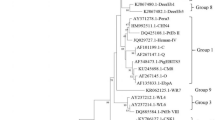
The SSU rRNA genes of Cryptosporidium spp. in all 23 microscopy-positive samples were successfully amplified with nested PCR. RFLP and DNA sequence analyses of the SSU rRNA gene fragments revealed the presence of three Cryptosporidium species: C. parvum (n = 15) on six farms, C. bovis (n = 4) on three farms, and C. andersoni (n = 4) on one farm (Table 1). With the exception of farm 2, only one Cryptosporidium species was detected on all the Cryptosporidium-positive farms (Table 1). The gp60 sequencing analysis showed that the 15 C. parvum isolates all belonged to subtype IIdA15G1.
Sequencing analyses of the tpi and gdh genes in G. duodenalis identified two assemblages: E (n = 25) on 10 farms and B (n = 4) on three farms (Table 1). Only farm 3 had both assemblages E and B.
Age distributions of Cryptosporidium and G. duodenalis
Cryptosporidium parvum was the most commonly identified Cryptosporidium species, responsible for 65.2% of all Cryptosporidium infections, and was only found in preweaned calves (Table 2). Cryptosporidium bovis and C. andersoni were found in preweaned calves and > 2-year-old cattle, respectively. In contrast, no Cryptosporidium-positive sample was identified in cattle aged between 3 months and 2 years.
Giardia duodenalis was only detected in preweaned and 3–11-month-old calves. Among these calves, G. duodenalis assemblage E was the dominant assemblage, responsible for 86.2% (25/29) of all Giardia-positive samples. In contrast, only four assemblage B infections (one subtype B1 and three subtype B2) were found in preweaned calves. Four mixed infections of Cryptosporidium and G. duodenalis were found in preweaned calves (Table 2).
Discussion
The overall infection rate for Cryptosporidium spp. was 1.68%, which is lower than those in Henan (13.0%, 276/2116) [12], Anhui (14.9%, 52/350), Jiangsu (20.7%, 251/1215), and Shanghai (12.5%, 55/440) [18]. The average infection rate for G. duodenalis was 2.12%, which is lower than those in Heilongjiang (5.2%, 42/814) [10] and Henan (7.2%, 128/1777) [13]. Oocysts shed at low intensity may be contributed to the low detecting rates of both parasites by microscopy. In general, it is difficult to explain the actual discrepancies in the prevalence of Cryptosporidium spp. and G. duodenalis among different studies because the infection rates are related to many factors, including the examination methods, age distributions of the animals, sample sizes, host health status at the time of sampling, the timing of specimen collection, and geo-ecological conditions. Nevertheless, the infection rates of both parasites were always higher in preweaned calves than in any other age group, which is consistent with previous observations [12]. The results of most previous studies, conducted in numerous countries, suggest that C. parvum is the predominant Cryptosporidium species in preweaned calves [12],[24],[13]. The results of most studies conducted in numerous countries, including Belgium, Denmark, Portugal, Spain, Sweden, Germany, UK, USA, Canada, Brazil, Australia, Rwanda, Egypt, Sri Lanka, and Malaysia, suggest that assemblage E is the predominant Giardia assemblage in cattle [13]. In general, assemblage B has been detected in only a small number of cattle in a few studies worldwide, including in Europe, Italy, Portugal, Uganda, New Zealand, and Canada [2],[30]. Assemblage B, one of the two major assemblages causing human giardiasis, has a broad host range, including cattle, sheep, pigs, horses, dogs, cats, and rabbits [2]. Although no strong evidence supporting the direct zoonotic transmission of G. duodenalis from animals to humans has been reported, case–control studies have shown that contact with farm animals increases the infection rate of human giardiasis [40],[41]. Considering how large the cattle industry is and the close contact that occurs between cattle and humans, assemblage B G. duodenalis identified in cattle may emerge as an important zoonotic pathogen in some areas of China.
Conclusion
In summary, C. parvum is the predominant species in preweaned calves in the study area, which clearly differs from the situation in Henan, Heilongjiang, and Shannxi Provinces. The presence of C. parvum subtype IIdA15G1, together with subtypes IIdA15G1 and IIdA19G1, previously found in rodents, ruminants, and humans, further confirms the dominance of the C. parvum IId subtypes in China. The dominant G. duodenalis assemblage, assemblage E, is similar to the dominant assemblages in other areas or countries.
References
**ao L, Ryan U: Molecular epidemiology. In Cryptosporidium and Cryptosporidiosis. 2nd edition. Edited by Fayer R, **ao L. Boca Raton: CRC Press and IWA Publishing; 2008:119–151
Feng Y, **ao L: Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011, 24: 110-140. 10.1128/CMR.00033-10.
Trout JM, Santín M: Livestock. In Cryptosporidium and Cryptosporidiosis. 2nd edition. Edited by Fayer R, **ao L. Boca Raton: CRC Press and IWA Publishing; 2008:451–483.
Santín M, Trout JM, Fayer R: A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet Parasitol. 2008, 155: 15-23. 10.1016/j.vetpar.2008.04.018.
Esteban E, Anderson BC: Cryptosporidium muris: prevalence, persistency, and detrimental effect on milk production in a drylot dairy. J Dairy Sci. 1995, 78: 1068-1072. 10.3168/jds.S0022-0302(95)76723-6.
Adam RD: Biology of Giardia lamblia . Clin Microbiol Rev. 2001, 14: 447-475. 10.1128/CMR.14.3.447-475.2001.
Liu A, Wang R, Li Y, Zhang L, Shu J, Zhang W, Feng YY, **ao LH, Ling H: Prevalence and distribution of Cryptosporidium spp. in dairy cattle in Heilongjiang Province, China. Parasitol Res. 2009, 105: 797-802. 10.1007/s00436-009-1457-2.
Wang R, Ma G, Zhao J, Lu Q, Wang H, Zhang L, Jian F, Ning C, **ao L: Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitol Int. 2011, 60: 1-4. 10.1016/j.parint.2010.09.002.
Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, Ning C, **ao L: Characteristics of Cryptosporidium transmission in preweaned dairy cattle in henan, China. J Clin Microbiol. 2011, 49: 1077-1082. 10.1128/JCM.02194-10.
Liu A, Zhang X, Zhang L, Wang R, Li X, Shu J, Zhang X, Shen Y, Zhang W, Ling H: Occurrence of bovine giardiasis and endemic genetic characterization of Giardia duodenalis isolates in Heilongjiang Province, in the Northeast of China. Parasitol Res. 2012, 111: 655-661. 10.1007/s00436-012-2883-0.
Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, Ling H, Liu A, Shen Y: Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China’s Heilongjiang Province. PLoS One. 2013, 8: e54857-10.1371/journal.pone.0054857.
Zhao G, Ren W, Gao M, Bian Q, Hu B, Cong M, Lin Q, Wang R, Qi M, Qi M, Zhu X, Zhang L: Genoty** Cryptosporidium andersoni in cattle in Shaanxi province, Northwestern China. PLoS One. 2013, 8: e60112-10.1371/journal.pone.0060112.
Wang H, Zhao G, Chen G, Jian F, Zhang S, Feng C, Wang R, Zhu J, Dong H, Hua J, Wang M, Zhang L: Multilocus genoty** of Giardia duodenalis in dairy cattle in Henan. China PLoS One. 2014, 9: e100453-10.1371/journal.pone.0100453.
Feng Y, Ortega Y, He G, Das P, Xu M, Zhang X, Fayer R, Gatei W, Cama V, **ao L: Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet Parasitol. 2007, 144: 1-9. 10.1016/j.vetpar.2006.10.001.
Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, **ao L: Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005, 43: 2805-2809. 10.1128/JCM.43.6.2805-2809.2005.
**ao L: Molecular epidemiology of cryptosporidiosis: An update. Exp Parasitol. 2010, 124: 80-89. 10.1016/j.exppara.2009.03.018.
Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E: Multilocus genoty** of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008, 38: 1523-1531. 10.1016/j.ijpara.2008.04.008.
Chen F, Huang K: Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle from farms in China. J Vet Sci. 2012, 13: 15-22. 10.4142/jvs.2012.13.1.15.
Geurden T, Vanderstichel R, Pohle H, Ehsan A, von Samson-Himmelstjerna G, Morgan ER, Camuset P, Capelli G, Vercruysse J, Claerebout E: A multicentre prevalence study in Europe on Giardia duodenalis in calves, with molecular identification and risk factor analysis. Vet Parasitol. 2012, 190: 383-390. 10.1016/j.vetpar.2012.06.039.
Minetti C, Taweenan W, Hogg R, Featherstone C, Randle N, Latham SM, Wastling JM: Occurrence and diversity of Giardia duodenalis assemblages in livestock in the UK. Transbound Emerg Dis in press.
**ao L, Fayer R: Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol. 2008, 38: 1239-1255. 10.1016/j.ijpara.2008.03.006.
Mi R, Wang X, Li C, Huang Y, Zhou P, Li Z, Lei M, Cai J, Chen Z: Prevalence and genetic characterization of Cryptosporidium in yaks in Qinghai Province of China. PLoS One. 2013, 8: e74985-10.1371/journal.pone.0074985.
Lv C, Zhang L, Wang R, Jian F, Zhang S, Ning C, Wang H, Feng C, Wang X, Ren X, Qi M, **ao L: Cryptosporidium spp. in wild, laboratory, and pet rodents in china: prevalence and molecular characterization. Appl Environ Microbiol. 2009, 75: 7692-7699. 10.1128/AEM.01386-09.
Li N, ** and subty** parasites in wastewater. PLoS Negl Trop Dis. 2012, 6: e1809-10.1371/journal.pntd.0001809.
Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, Liu L, Feng Y, **ao L: Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013, 51: 557-563. 10.1128/JCM.02758-12.
Insulander M, Silverlås C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B: Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect. 2013, 141: 1009-1020. 10.1017/S0950268812001665.
Amer S, Zidan S, Feng Y, Adamu H, Li N, **ao L: Identity and public health potential of Cryptosporidium spp. in water buffalo calves in Egypt. Vet Parasitol. 2013, 191: 123-127. 10.1016/j.vetpar.2012.08.015.
Iqbal A, Lim YA, Surin J, Sim BL: High diversity of Cryptosporidium subgenotypes identified in Malaysian HIV/AIDS individuals targeting gp60 gene. PLoS One. 2012, 7: e31139-10.1371/journal.pone.0031139.
Imre K, Luca C, Costache M, Sala C, Morar A, Morariu S, Ilie MS, Imre M, Dărăbuş G: Zoonotic Cryptosporidium parvum in Romanian newborn lambs (Ovis aries). Vet Parasitol. 2013, 191: 119-122. 10.1016/j.vetpar.2012.08.020.
Budu-Amoako E, Greenwood SJ, Dixon BR, Barkema HW, McClure JT: Giardia and Cryptosporidium on dairy farms and the role these farms may play in contaminating water sources in Prince Edward Island, Canada. J Vet Intern Med. 2012, 26: 668-673. 10.1111/j.1939-1676.2012.00930.x.
Mark-Carew MP, Wade SE, Chang YF, Schaaf S, Mohammed HO: Prevalence of Giardia duodenalis assemblages among dairy herds in the New York City Watershed. Vet Parasitol. 2012, 185: 151-157. 10.1016/j.vetpar.2011.09.030.
Muhid A, Robertson I, Ng J, Yang R, Ryan U: Prevalence of Giardia spp. infection in pre-weaned and weaned calves in relation to management factors. Vet J. 2012, 191: 135-137. 10.1016/j.tvjl.2011.01.007.
Silva FM P e, Lopes RS, Araújo JP: Genetic characterisation of Giardia duodenalis in dairy cattle in Brazil. Folia Parasitol (Praha). 2012, 59: 15-20. 10.14411/fp.2012.003.
Santin M, Dargatz D, Fayer R: Prevalence of Giardia duodenalis assemblages in weaned cattle on cow-calf operations in the United States. Vet Parasitol. 2012, 183: 231-236. 10.1016/j.vetpar.2011.07.042.
Amer SE: Genotypic and phylogenetic characterization of Giardia intestinalis from human and dairy cattle in Kafr El Sheikh Governorate, Egypt. J Egypt Soc Parasitol. 2013, 43: 133-146.
Fava NM, Soares RM, Scalia LA, Kalapothakis E, Pena IF, Vieira CU, Faria ES, Cunha MJ, Couto TR, Cury MC: Performance of glutamate dehydrogenase and triose phosphate isomerase genes in the analysis of genotypic variability of isolates of Giardia duodenalis from livestocks. Biomed Res Int. 2013, 2013: 875048-10.1155/2013/875048.
Gillhuber J, Pallant L, Ash A, Thompson RC, Pfister K, Scheuerle MC: Molecular identification of zoonotic and livestock-specific Giardia-species in faecal samples of calves in Southern Germany. Parasitol Vectors. 2013, 6: 346-10.1186/1756-3305-6-346.
Abeywardena H, Jex AR, Koehler AV, Rajapakse RP, Udayawarna K, Haydon SR, Stevens MA, Gasser RB: First molecular characterization of Cryptosporidium and Giardia from bovines (Bos taurus and Bubalus bubalis) in Sri Lanka: unexpected absence of C. parvum from pre-weaned calves. Parasitol Vectors. 2014, 7: 75-10.1186/1756-3305-7-75.
Hogan JN, Miller WA, Cranfield MR, Ramer J, Hassell J, Noheri JB, Conrad PA, Gilardi KV: Giardia in mountain gorillas (Gorilla beringei beringei), forest buffalo (Syncerus caffer), and domestic cattle in Volcanoes National Park, Rwanda. J Wildl Dis. 2014, 50: 21-30. 10.7589/2012-09-229.
Hoque ME, Hope VT, Kjellstrom T, Scragg R, Lay-Yee R: Risk of giardiasis in Aucklanders: a case–control study. Int J Infect Dis. 2002, 6: 191-197. 10.1016/S1201-9712(02)90110-4.
Hoque ME, Hope VT, Scragg R, Kjellstrom T: Children at risk of giardiasis in Auckland: a case–control analysis. Epidemiol Infect. 2003, 131: 655-662. 10.1017/S0950268803008598.
Acknowledgements
This study was supported, in part, by the National Natural Science Foundation of China (U1204328 and 31302079), the Specialized Research Fund for the Doctoral Program of Higher Education (20124105120003), and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-37).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LXZ and RJW designed the experiments; JYH, DYY, and MQ performed the study; JFZ and JQL analyzed the data; KS and MW contributed reagents/materials/analysis tools; RJW, LXZ, and JYH wrote the paper. All authors have read and approved the final version of the manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Huang, J., Yue, D., Qi, M. et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalisin dairy cattle in Ningxia, northwestern China. BMC Vet Res 10, 292 (2014). https://doi.org/10.1186/s12917-014-0292-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-014-0292-6