Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) is a common chronic liver illness with a genetically heterogeneous background that can be accompanied by considerable morbidity and attendant health care costs. The pathogenesis and progression of NAFLD is complex with many unanswered questions. We conducted genome-wide association studies (GWASs) using both adult and pediatric participants from the Electronic Medical Records and Genomics (eMERGE) Network to identify novel genetic contributors to this condition.
Methods
First, a natural language processing (NLP) algorithm was developed, tested, and deployed at each site to identify 1106 NAFLD cases and 8571 controls and histological data from liver tissue in 235 available participants. These include 1242 pediatric participants (396 cases, 846 controls). The algorithm included billing codes, text queries, laboratory values, and medication records. Next, GWASs were performed on NAFLD cases and controls and case-only analyses using histologic scores and liver function tests adjusting for age, sex, site, ancestry, PC, and body mass index (BMI).
Results
Consistent with previous results, a robust association was detected for the PNPLA3 gene cluster in participants with European ancestry. At the PNPLA3-SAMM50 region, three SNPs, rs738409, rs738408, and rs3747207, showed strongest association (best SNP rs738409 p = 1.70 × 10− 20). This effect was consistent in both pediatric (p = 9.92 × 10− 6) and adult (p = 9.73 × 10− 15) cohorts. Additionally, this variant was also associated with disease severity and NAFLD Activity Score (NAS) (p = 3.94 × 10− 8, beta = 0.85). PheWAS analysis link this locus to a spectrum of liver diseases beyond NAFLD with a novel negative correlation with gout (p = 1.09 × 10− 4). We also identified novel loci for NAFLD disease severity, including one novel locus for NAS score near IL17RA (rs5748926, p = 3.80 × 10− 8), and another near ZFP90-CDH1 for fibrosis (rs698718, p = 2.74 × 10− 11). Post-GWAS and gene-based analyses identified more than 300 genes that were used for functional and pathway enrichment analyses.
Conclusions
In summary, this study demonstrates clear confirmation of a previously described NAFLD risk locus and several novel associations. Further collaborative studies including an ethnically diverse population with well-characterized liver histologic features of NAFLD are needed to further validate the novel findings.
Similar content being viewed by others
Background
Nonalcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases, found in 17–30% of the population in Western countries [1]. NAFLD, defined as greater than 5% fatty acid content of liver by weight, includes not only simple and benign steatosis but also the more serious nonalcoholic steatohepatitis (NASH), which may progress to cirrhosis and liver failure in 8 to 26% of adults with NASH [2]. NASH is defined histologically by the presence of macrovesicular steatosis, lobular inflammation, and hepatocellular ballooning. The pathology is often indistinguishable from alcoholic fatty liver disease; therefore, the diagnosis can only be made in the absence of significant alcohol use [3]. NAFLD is now recognized as a common metabolic disorder globally as a result of ongoing obesity pandemic. It also increases risk of adverse long-term consequences including death from liver cirrhosis and cardiovascular disease. In fact, NASH is now the second most common indication for liver transplantation in the USA after chronic hepatitis C [4].
Growing evidence has shown that NAFLD can also occur in 10–20% of non-obese population, most often in association with central adiposity, recent weight gain, dietary factors, or genetic risk alleles [5]. In East Asian countries, for example, the incidence and prevalence of NAFLD are increasing with time despite lower rates of obesity compared to Western countries [6]. Hence, it is important to identify the natural course of NAFLD and the contributing factors for the development and maintenance or regression of this disease. The underlying etiology is believed to be multifactorial with a substantial genetic component. The heritability estimates of NAFLD generally range from 20 to 70%, depending on the study design, ethnicity, and the methodology used [7]. Likewise, for indices of disease severity, the heritability estimates in a twin study for hepatic steatosis was 0.52 (based on MRI proton-density fat fraction) and for liver fibrosis (based on liver stiffness) 0.5 [8]. In addition, heritability risk for NAFLD may be independent of body mass index heritability. For example, family studies show that while fatty liver can be present in 17% of siblings and 37% of parents of overweight children without NAFLD, it was significantly more common in siblings (59%) and parents (78%) of children with NAFLD [9]. To date, several genome-wide association studies (GWAS) have been published for this condition mainly in adult cohorts [10,11,12]. One of the established effects is in the PNPLA3 (patatin-like phospholipase domain–containing 3) gene with consistent results across studies in which the rs738409 C>G variant (resulting in an amino acid substitution of methionine for isoleucine at position 148 (I148M)) is strongly associated with this trait. The PNPLA3 protein exerts lipase activity and plays a role in the hydrolysis of glycerolipids, with maximum enzymatic activity against triglycerides, diacylglycerol, and monacylglycerol [13]. Structural modeling suggests that this substitution may occlude access of substrates to the catalytic dyad [14]. However, the exact underlying mechanisms remain unclear.
The electronic medical record (EMR) is a rich source of clinical information. Natural language processing (NLP) techniques have demonstrated successes within the clinical domain and have been tested for transferability to another institution [15]. The electronic MEdical Records and GEnomics (eMERGE) Network, founded in 2007, is a consortium of multiple adult and pediatric institutions developed to explore the utility of DNA biorepositories linked to EMRs as well as establishing and validating specific algorithms with and without NLP for many common phenotypes [16]. In this study, we investigated the genetic variants associated with NAFLD/NASH in children and adults using phenotypic measures extracted from medical records in a collection of already genotyped samples from more than 80,000 eMERGE participants to replicate prior studies and identify additional genetic loci.
Methods
Study participants and phenotype
Data for this study were collected from the eMERGE Network [17]. Protocols for this study were approved by the Institutional Review Boards (IRBs) at the institutions where participants were recruited; all included participants provided written informed consent prior for inclusion in the study. The population comprised 9677 unrelated European ancestry participants (1106 cases and 8571 controls). A natural language processing (NLP) algorithm was deployed in each site to identify NAFLD cases and controls. These include logic concepts using billing codes, laboratory values, text queries, and medication records to identify true cases and controls at each site. A rules-based NLP algorithm was developed using structured and unstructured data from Cincinnati Children’s Hospital and Medical Center (CCHMC) and secondarily validated in Children’s Hospital of Philadelphia (CHOP) with high precision. The eMERGE protocol includes development of an algorithm at a primary site and the implementation and validation at a secondary site. The secondary site serves as a testing ground for the purposes of mitigating overfitting concerns and ensuring portability. Expert validation includes manual chart review at each site by a physician for both cases and controls. After obtaining a validated positive predictive value of 95% for cases and controls at both the primary (CCHMC) and secondary sites (CHOP), the algorithm has been implemented across network. The exclusion and inclusion criteria for NAFLD were derived according to recommendation from the American Association for the Study of Liver Diseases (AASLD) practical guideline for NAFLD [18]. Case inclusion and exclusion criteria, list of excluded medications, and the number of participants per eMERGE site can be found in Additional file 1: Table S1. We processed the pathology and radiology reports from encounters with diagnosis codes by searching with regular expressions for specific related terms as shown in Additional file 1: Table S1. The NegEx multilingual lexicon was used to assess positive and negative condition for each term [19]. In addition, NAFLD disease severity was assessed based on available liver enzyme and histopathologic grade using the NAFLD Activity Score (NAS). NAS score is a standard method used to score NAFLD disease activity and originally has been developed as a tool to measure disease prognosis and changes in NAFLD during therapeutic trials [20]. The NAS is derived from an unweighted sum of scores of liver steatosis (0–3), lobular inflammation (0–3), and hepatocellular ballooning (0–2), ranging between 0 and 8. Coexistent fibrosis also has a separate scoring range of 0–4. This consists of no fibrosis (0), perisinusoidal or periportal (1), portal (2), bridging fibrosis (3), and cirrhosis (4). The NAS classification scoring system is shown in Additional file 1: Table S1. We obtained these values from pathology reports using NLP processing for 235 of our NAFLD case participants. In addition, for each case, the highest level of liver enzyme values for aspartate aminotransferase (AST U/L) and alanine aminotransferase (ALT U/L) was obtained for association testing.
Genoty** and imputation
Genetic data for the eMERGE Network is available from the coordinating center and can be accessed through dbGAP (phs000888.v1.p1) which is annually updated. High-throughput SNP genoty** was carried out previously in each contributing medical center. A series of standard quality control (QC) measures has been applied before and after imputation. These measures have been developed by the eMERGE Genomics Workgroup [21, 22]. The standard QC process included sample call rates, sample relatedness, and population stratification, sex inconsistency as well as marker quality (i.e., marker call rate, minor allele frequency (MAF), and Hardy-Weinberg equilibrium (HWE). In this study, all analyses were limited to participants with call rates > 98%, SNPs with call rates > 99%, and SNPs with MAF > 1% and HWE p > 0.0001 in controls. The details of imputation process and principal component (PC) analyses have been included in Additional file 2 [23–25].
Statistical analyses
Logistic (case-control) and quantitative linear (case-only) regression analyses were performed using an additive genetic model adjusting for 10 medical centers; PCs 1, 2, and 3; sex; and age. In addition, since NAFLD is closely linked to obesity, we included the most recent BMI for each subject as another covariate and remove all missing participants from analyses. Traditionally absolute BMI (kg/m2) is used for adults, while age- and sex-specific BMI-z scores and percentiles apply in children and adolescents to account for their continuing growth. In combined analyses, we therefore transformed all BMI into 6 classes: underweight (< 18.5 or < 5th percentile), normal (18.50–24.99 or 5th to < 85th percentile), overweight (25.00–29.99 or 85th to < 95th percentile), and obese: class 1 (30–34.99 or 95th to < 120% of the 95th percentile), class 2 (35–39.99 or 120% to < 140% of the 95th percentile), and class 3 (≥ 40 or ≥ 140% of the 95th percentile). The percentage of BMI ≥ 95% in pediatrics participants was estimated using the CDC-based online resource [27]. The distribution of participants that we received from the network also varied per site (see Additional file 1: Table S1); we therefore adjust for 10 study sites. Adjusting for too many covariates may sometimes cause the standard logistic regression to fail to converge especially for less-frequent variants. Firth’s penalized likelihood approach, available in second generation of PLINK, is a method of addressing issues of separation and bias of the parameter estimates in which we used in regression analyses when necessary [26]. For liver enzymes (AST, ALT), we used the highest value U/L per subject. All quantitative phenotypes including liver enzymes and NAS score were standardized to mean of zero and variance 1 using PLINK.
Further conditional analyses and pairwise SNP × SNP interactions were also performed using “epistasis” option in PLINK. In this study, we only analyzed the pairwise interaction effect of one known SNP (rs738409) in PNPLA3 gene against the genome. The slower “—epistasis” command was used to test for epistasis using logistic regression which is the most accurate test to define SNP × SNP interactions [26]. Interactions were excluded if two SNPs were located within 1 Mb of each other to avoid spurious evidence of interaction due to linkage disequilibrium (LD). Narrow-sense heritability was also estimated using an SNP-based approach available in the GCTA program [28] which evaluates the proportion of phenotypic variance explained by all SNPs. Briefly, the GCTA analysis consists of two steps. First, all SNPs are used to calculate the genetic relationship matrix (GRM) among participants using the observed low-level genetic similarity in SNP data from individuals who are not directly related. This measure is then used as a predictor in a mixed linear model with a trait as the response to estimate h2 [28]. The weighted genetic risk score (GRS) was also calculated using PLINK-score function by multiplying each β-coefficient of highly significant SNPs with the number of corresponding risk alleles (0, 1, or 2) and then summing the products ( [26]. For known variants, β-coefficients were obtained from the GWAS catalog [29]. The performance of the obtained GRS score for disease diagnosis and prediction accuracy were evaluated using receiver operating characteristic (ROC) curve, using MedCalc software [30]. Finally, to estimate the level of heterogeneity between pediatrics and adult cohorts, Cochran’s Q test statistics was applied using PLINK2 [26].
PheWAS analyses
A phenome-wide association study (PheWAS) was also performed in order to evaluate pleotropic effects of the known GWAS variant (rs738409) as well as other novel effects in this study with any other trait in children or adults. The trait definition in PheWAS approach is mainly based on billing International Classification of Diseases (ICD) codes; therefore, it is less conservative. The detail of this approach has been described previously [31]. We used the PheWAS package in R version 3.5.1 [32]. Briefly, in the PheWAS process, first the ICD-9 codes are collapsed into PheWAS codes according to PheWAS map [32]. Then, cases and controls are determined according to the code under study. In these analyses, a case was defined as having at least two occurrences of the PheWAS code on different days and the controls with no instances. Additionally, we used a threshold of at least 20 cases for the code to be used in the model. Next, for each PheWAS code, a logistic regression model was created and adjusted for age, sex, BMI, site of genoty**, and PCs similar to GWAS study. A false discovery rate (FDR) of 0.05 using the Benjamini–Hochberg procedure implemented in PheWAS was then used to correct the threshold for multiple hypotheses testing.
Post-GWAS analyses and data visualization
The details of post-GWAS analyses including functional annotation, prioritization, and interpretation of GWAS results based on functional map** are included in Additional file 2 [33–41].
Power analyses
We used QUANTO for power calculation of case-only and case-control GWAS analyses [42]. For quantitative NAS-score analysis with 235 participants, given the mean and standard deviation of our continuous variable, i.e., NAS score (mean 3.78, SD 1.76) (see Table 1), we tested the power assuming an additive genetic model. For variants with minor allele frequency above 0.2 and effect size (βG) of at least 0.5, this sample size will have > 0.80 power to identify the association at an alpha level of 0.05. Of note, almost all of our top genetic associations for NAS score or fibrosis had minor allele frequency above 0.2. In case-control GWAS analyses with 1106 cases and 8571 controls, we had more than 90% power to detect effects for all variants with MAF > 0.01 under an additive model.
Results
The results reported below consist of overall NAFLD case-control GWAS and four additional case-only GWA quantitative studies for NAS score, fibrosis, and AST and ALT liver enzymes.
NAFLD case-control GWAS
Table 2 shows demographic characteristics of patients and controls included in this study. The mean age was 63.5 (±16.86 SD) for adult participants (N = 8435) and 13.05 (±5.41 SD) for pediatric participants (N = 1242). The number of participants per site is included in Additional file 1: Table S1. In this study, 47% of pediatric participants and 42% of adults were males. A total of 9677 unrelated European ancestry participants (1106 cases and 8571 controls) and 7,263,501 autosomal variants were evaluated for this GWAS analysis.
Associations of previously reported SNPs
Consistent with previous reports, we identified strong genetic signals at the PNPLA3 locus at 22q13. Figure 1a shows a Manhattan plot with one main peak located on chromosome 22 that was associated with NAFLD. The Q–Q plot of this GWAS is also shown in Fig. 1b. The overall low inflation rate of λ = 1.001 indicated no major population stratification. At the PNPLA3-SAMM50 region, three proxy SNPs (r2 > 0.95), rs738409, rs738408, and rs3747207, located in the PNPLA3 gene showed the strongest associations (best SNP rs738409 p = 1.70 × 10− 20, OR = 1.79 (95% CI = 1.58–2.02)) (Table 3, Fig. 2a). This effect was consistent in both pediatric (p = 9.92 × 10− 6, OR = 1.76 (95% CI = 1.37–2.27)) and adult (9.73 × 10− 15, OR = 1.79 (95% CI = 1.55–2.08)) cohorts and with no evidence of heterogeneity (Cochran’s Q = 0.78, I2 = 0). Consistent with previous results, another coding variant rs2294918 (E434K) in PNPLA3 gene was associated at a weaker level (p = 1.90 × 10− 5). The SNPs with the most significant evidence for association are summarized in Table 3, and all results with p < 10− 5 are included in Additional file 1: Table S2.
a, b Manhattan plot (a) and Q–Q plot (b) of genome-wide markers for NAFLD in European ancestry (1106 cases and 8571 controls). A total of 1106 cases of NAFLD and 8571 controls were analyzed after quality control. Logistic regression analysis was performed for 7,261,527 variants with MAF > 1% assuming an additive genetic model, adjusted for age, sex, BMI, genoty** site, and genetic ancestry (principal components 1 through 3). Results are plotted as –log10 p values on the y-axis by position in chromosome (x-axis) (NCBI build 37)
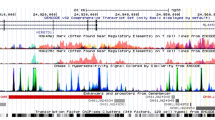
a–c LocusZoom plot of the associations signals in three previously known regions for NAFLD. a Confirmation at 22q13 for PNPLA3. SNP rs738409 is a missense variation (I148M) in PNPLA3 produced the best effect (p = 1.70 × 10− 20). b Detected signal at 19p12 (GATAD2A, NCAN, TM6SF2) region. The best marker in this study was rs56408111 (p = 5.26 × 10− 6). The linkage disequilibrium (LD) between rs56408111 and previously known SNP rs4808199 was r2 = 0.24, D’ = 0.74. c Detected signal at 8q24 (TRIB1) genetic region. The best marker in this study (rs2980888) is shown (see also Additional file 1: Table S2). Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the most significant variant are color-coded to reflect their LD with the index SNP (taken from pairwise r2 values from the HapMap CEU database, www.hapmap.org). Regional plots were generated using LocusZoom (http://csg.sph.umich.edu/locuszoom)
Apart from the main effect at the PNPLA3 locus, previous genetic studies identified several minor effects at other chromosomal loci, including GCKR at 2p23, and GATAD2A, NCAN, and TM6SF2 at 19p12 [43, 44]. We examined whether or not the associations were reproduced in our cohorts by extracting genotype information of SNP markers corresponding to these loci. None of these effects reached genome-wide significance. In our pediatric cohorts, the association of rs1260326 and rs780094 in GCKR was borderline significant (p = 0.006, OR = 1.40, 95% CI 1.1–1.78). However, the association was lost when examined in the adult cohort (see Additional file 1: Table S3). At the 19p12 region (GATAD2A, NCAN, TM6SF2), the association with known SNP rs4808199 was also detected using all cohorts (p = 0.004, OR = 1.22, 95% CI 1.06–1.40)). Of note, the known TM6SF2 missense variant rs58542926 (E167K) produced a p = 0.03 (OR = 1.23, 95% CI 1.01–1.52) in our cohort. In this region, we also found other unreported downstream markers with stronger associations (best effect for SNP rs56408111 p = 5.26 × 10− 6) (see Additional file 1: Table S2; Fig. 2b). The observed effect for rs4808199 or rs58542926 disappeared after conditioning on rs56408111 (p = 0.71, p = 0.17 respectively) suggesting that the association in this region mostly derives from rs56408111. The LD between these two known markers (rs4808199, rs58542926) and the best variant in this study, rs56408111, was modest (r2 = 0.25, r2 = 0.40 respectively).
We also confirmed an effect at 8q24 near the TRIB1 gene that previously associated with NAFLD in the Japanese population [45]. In their population, rs2954021 produced p = 4.5 × 10− 5. In our European ancestry population and for the first time, this variant as well as a cluster of variants nearby was associated with NAFLD with the best marker rs2980888 (p = 5.98 × 10− 7, OR = 1.36 95% CI = 1.20–1.53) (see Table 3, Fig. 2c). Conditional analyses suggest that rs2980888 is the most informative variant in this region in European ancestry. These two markers resided in one risk haplotype in European ancestry with (r2 = 0.45, D’ = 0.97) (Fig. 2c).
Controlling for the main effects at PNPLA3
We used logistic regression models conditioned on the main effect at PNPLA3 as well as testing for epistatic interaction between the known SNP at PNPLA3 rs738409 and the rest of genome. For conditional analysis, the genotype data of rs738409 in dosage format (0, 1, 2) was included as another covariate in addition to age, sex, PCs, BMI, and sites of genoty**. While this variant controlled all effects at PNPLA3 indicating no other independent effects at this locus, no major changes have been detected in other loci. Next, in a separate analysis, the epistatic effect of the known SNP rs738409 with the rest of the genome was evaluated (see “Methods”). Several suggestive results were detected across the genome with only one effect at 16p12 that passed the significance threshold of p < 0.0001 (SNP rs2188761, case-only Pepistasis = 2.47 × 10− 7, case-control Pepistasis = 7.32 × 10− 6, OR of interaction = 1.50). Several proxy markers in this region (16p12) such as rs7499477, rs2188760, and rs6497497 (r2 > 0.95 with rs2188761) also interact with rs738409 in PNPLA3. In addition, this novel epistatic effect was consistent in both pediatrics and adult cohorts (OR of interaction = 1.57 and 1.43 for pediatrics and adults, respectively). As mentioned above, all these markers had passed QC and were in HWE. However, none of these markers at the 16p12 region were genome-wide significant in GWAS analyses (0.1 > p > 0.02).
Because NAFLD is closely linked to obesity, we also explored the specific SNP × SNP interaction of the major obesity locus, FTO (rs1421085) and PNPLA3 (rs738409). We did not find any significant SNP × SNP interaction (p = 0.72). Of note, the GWAS effect for FTO (rs1421085) in this study was p = 0.25 after controlling for BMI. However, by relaxing the model and removing the BMI as a covariate, this effect in FTO increased in significance (p = 9.26 × 10− 6).
Case-only GWA studies
Impact of SNPs on the severity of NAFLD
We next investigated the associations of the SNPs with NAFLD disease severity based on available histopathologic grade, namely, NAS and liver enzymes (see “Methods”). Because liver biopsy usually is not indicated for NAFLD diagnosis, we were able to identify and score only 235 participants using EMR data from the total of 1106 NAFLD cases that includes 107 pediatric and 128 adult cases (Table 1). Liver function tests (AST U/L, ALT U/L) were available for 1075 of case participants. In addition to the main case-control study, for disease severity index, we performed GWAS for each quantitative trait (NAS score, fibrosis, ALT, AST) using linear regression method adjusting for age, sex, BMI, PCs, and site of genoty**. These case-only analyses showed several loci with significant associations. The SNPs with the most significant evidence are summarized in Table 3 and all results with p < 10− 5 are provided in Additional file 1: Table S2. Consistent with previous reports, index SNP rs738409 at PNPLA3 showed a significant association with disease severity NAS score (p = 3.94 × 10− 8, beta = 0.85) (Table 3). Indeed, if we consider a binary outcome in which NAS score ≥ 5 as case versus the remaining cases as control (79 cases versus 156 controls), an OR = 2.72, 95% OR (1.83–4.04), and p = 4.27 × 10− 7 can be obtained for this marker. As shown in Table 1, 34% of our participants had a NAS score ≥ 5. Figure 3 also shows the mean of NAS score and fibrosis together (0–12) stratified by PNPLA3 index SNP rs738409-genotype (GG/GC/CC) in which a beta of 1.07 (SE = 0.20) can be obtained. This is almost equal to one unit increase in NAS scores per risk allele.
a Means and standard deviations of NAS and fibrosis score (0–12) stratified by genotype of rs738409 at PNPLA3 in 235 NAFLD cases. The results are plotted as the sum of NAS and fibrosis score (0–12) (y-axis) against the three genotypes of rs738409 C>G polymorphism (x-axis). The results are further sub-divided by age groups (pediatrics, adult, and all). Results for IL17RA (b) and ZFP90 (c) also are shown
Apart from the main effect at the PNPLA3 locus that also was associated with disease activity, previous genetic studies also identified several effects for histologic NAS score, fibrosis, and liver enzyme in NAFLD cases [10, 46]. In particular, Chalasani et al. evaluated 236 well-characterized NAFLD European ancestry female cases using 324,623 SNP markers for the histologic traits. After extracting genotype information of SNP markers corresponding to these loci, none of these effects reached genome-wide significance in our cohort. However, an effect for SNP rs1227756 at COL13A1 was associated (p = 0.008) with the NAS score (Additional file 1: Table S3). Another published effect was at chromosome 8 for SNP rs2645424 near FDFT1 for NAS score; however, a subsequent study failed to confirm it [47]. While association with this marker was weak in our cohort (best p = 0.15 for fibrosis), several nearby markers in this region were suggestively associated including eQTL variant rs1908814 (best p = 1.49 × 10− 4) for the same trait but with low LD with the previously published marker (r2 = 0.01) (see Additional file 1: Table S3). A recent study also identified an association of a splice variant in HSD17B13 (rs72613567:TA insertion) with reduced risk of NAFLD (p = 1.3 × 10− 5) [ In summary, we report genome-wide significant loci associated with NAFLD and disease severity index in a GWAS analysis of 9677 European ancestry individuals from 10 eMERGE study cohorts. Apart from the PNPLA3 effect, the GWAS implicates IL17RA and other biologically informative genes as important contributors to disease severity of NAFLD. The results also highlight strong overlap of PNPLA3 in the genetics of NAFLD and other liver pathologies and metabolic traits in the population, indicating a spectrum of conditions.Conclusion
Availability of data and materials
Genetic data for the eMERGE Network is available from the coordinating center and can be accessed through dbGAP (phs000888.v1.p1).
Abbreviations
- AASLD:
-
American Association for the Study of Liver Diseases
- ACSM5 :
-
Acyl-CoA Synthetase Medium chain family member 5 (gene)
- ALT:
-
Alanine aminotransferase
- AST:
-
Aspartate aminotransferase
- AUC:
-
Area under the ROC curve
- BMI:
-
Body mass index
- CADD:
-
Combined Annotation-Dependent Depletion
- CDH1 :
-
Cadherin 1 (gene)
- CEPBA :
-
CCAAT/enhancer binding protein alpha (C/EBP) (gene)
- COL13A1 :
-
Collagen type XIII alpha 1 chain (gene)
- eMERGE:
-
The electronic MEdical Records and GEnomics Network
- EMR:
-
Electronic medical record
- FABP1 :
-
Fatty acid-binding protein 1 (gene)
- FDR:
-
False discovery rate
- FLI1 :
-
Fli-1 proto-oncogene ETS transcription factor (gene)
- FTO :
-
FTO alpha-ketoglutarate dependent dioxygenase (gene)
- FUMA:
-
Functional map** and annotation of genome-wide association studies
- GATAD2A :
-
GATA zinc finger domain containing 2A (gene)
- GCKR :
-
Glucokinase regulator (gene)
- GRS:
-
Genetic risk score
- GTEX:
-
Genotype-Tissue Expression
- GWAS:
-
Genome-wide association study
- HRC:
-
Haplotype Reference Consortium
- HSD17B13 :
-
Hydroxysteroid 17-beta dehydrogenase 13 (gene)
- HWE:
-
Hardy-Weinberg equilibrium
- ICD:
-
International Classification of Diseases
- IL17RA :
-
Interleukin 17 receptor A (gene)
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MIS:
-
Michigan Imputation Server
- MSigDB:
-
Molecular Signatures Database
- NAFLD:
-
Nonalcoholic fatty liver disease
- NAS:
-
NAFLD Activity Score
- NASH:
-
Nonalcoholic steatohepatitis
- NCAN :
-
Neurocan (gene)
- NFIL3 :
-
Nuclear factor interleukin 3 regulated (gene)
- NLP:
-
Natural language processing
- PARVB :
-
Parvin beta (gene)
- PC:
-
Principal component
- PheWAS:
-
Phenome-wide association study
- PNPLA3 :
-
Patatin-like phospholipase domain–containing 3 (gene)
- PPARG :
-
Peroxisome proliferator-activated receptor gamma (gene)
- QC:
-
Quality control
- RELI:
-
Regulatory Element Locus Intersection
- ROC:
-
Receiver operating characteristic curve
- SAMM50 :
-
SAMM50 sorting and assembly machinery component (gene)
- SDC1 :
-
Syndecan 1 (gene)
- SPI1 :
-
Spi-1 proto-oncogene (gene)
- TM6SF2 :
-
Transmembrane 6 superfamily member 2 (gene)
- TRIB1 :
-
Tribbles pseudokinase 1 (gene)
- XDH :
-
Xanthine dehydrogenase (gene)
- ZFP90 :
-
ZFP90 zinc finger protein (gene)
References
Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–90.
Masuoka HC, Chalasani N. Nonalcoholic fatty liver disease: an emerging threat to obese and diabetic individuals. Ann N Y Acad Sci. 2013;1281:106–22.
Sanyal AJ, Brunt EM, Kleiner DE, Kowdley KV, Chalasani N, Lavine JE, et al. Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatol. 2011;54:344–53.
Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330–44.
Younes R, Bugianesi E. NASH in lean individuals. Semin Liver Dis. 2019;39:86–95.
Kim HJ, Kim HJ, Lee KE, Kim DJ, Kim SK, Ahn CW, et al. Metabolic significance of nonalcoholic fatty liver disease in nonobese, nondiabetic adults. Arch Intern Med. 2004;164:2169–75.
Sookoian S, Pirola CJ. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol. 2017;23:1–12.
Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, et al. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149:1784–93.
Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585–92.
Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, Nonalcoholic Steatohepatitis Clinical Research Network, et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology. 2010;139:1567–76 1576.e1–6.
Kawaguchi T, Shima T, Mizuno M, Mitsumoto Y, Umemura A, Kanbara Y, et al. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS One. 2018;1:e0185490.
Kitamoto T, Kitamoto A, Yoneda M, Hyogo H, Ochi H, Nakamura T, et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet. 2013;132:783–92.
Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;28:37085–93.
He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;26:6706–15.
Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7:41.
McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genet. 2011;4:13.
eMERGE Network. https://emerge.mc.vanderbilt.edu. 2007. Accessed 15 May 2018.
Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–57.
Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10.
Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–21.
Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet. 2011;68:1–19.
Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC, et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35:887–98.
Stanaway IB, Hall TO, Rosenthal EA, Palmer M, Naranbhai V, Knevel R, et al. The eMERGE genotype set of 83,717 subjects imputed to ~40 million variants genome wide and association with the herpes zoster medical record phenotype. Genet Epidemiol. 2019;43:63–81.
Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
PediTools, Clinical tools for pediatric providers. 2012. https://peditools.org. Accessed 20 Aug 2018.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog). Nucleic Acids Res. 2017;4:D896–901.
MedCalc Statistical Software version 18.6 (MedCalc Software bvba, Ostend, Belgium). 2018. http://www.medcalc.org. Accessed 10 Aug 2018.
Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10.
Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–6.
Watanabe E, Taskesen A, Bochoven v, Posthuma D. Functional map** and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. 2015;13:311–9.
Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4.
Zhbannikov IY, Arbeev K, Ukraintseva S, Yashin AI. haploR: an R package for querying web-based annotation tools. Version 2. F1000Res. 2017;6:97.
De Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Harley JB, Chen X, Pujato M, Miller D, Maddox A, Forney C, et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet. 2018;50:699–707.
Fang H, Knezevic B, Burnham KL, Knight JC. XGR software for enhanced interpretation of genomic summary data, illustrated by application to immunological traits. Genome Med. 2016;8:129.
Golden Helix GenomeBrowse1 visualization tool (SNP and variation suite” Version 8.8.1). Bozeman, MT: Golden Helix, Inc. (http://www.goldenhelix.com)). Accessed 5 Aug 2018.
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7.
Gauderman WJ, Morrison, JM. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. 2006; http://hydra.usc.edu/gxe.
Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324.
Wang X, Liu Z, Wang K, Wang Z, Sun X, Zhong L, et al. Additive effects of the risk alleles of PNPLA3 and TM6SF2 on non-alcoholic fatty liver disease (NAFLD) in a Chinese population. Front Genet. 2016;7:140.
Kitamoto A, Kitamoto T, Nakamura T, Ogawa Y, Yoneda M, Hyogo H, et al. Association of polymorphisms in GCKR and TRIB1 with nonalcoholic fatty liver disease and metabolic syndrome traits. Endocr J. 2014;61:683–9.
Larrieta-Carrasco E, Flores YN, Macías-Kauffer LR, Ramírez-Palacios P, Quiterio M, Ramírez-Salazar EG, et al. Genetic variants in COL13A1, ADIPOQ and SAMM50, in addition to the PNPLA3 gene, confer susceptibility to elevated transaminase levels in an admixed Mexican population. Exp Mol Pathol. 2018;104:50–8.
Stättermayer AF, Rutter K, Beinhardt S, Wrba F, Scherzer TM, Strasser M, et al. Role of FDFT1 polymorphism for fibrosis progression in patients with chronic hepatitis C. Liver Int. 2014;34:388–95.
Abul-Husn NS, Cheng X, Li AH, **n Y, Schurmann C, Stevis P, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–106.
Ma Y, Belyaeva OV, Brown PM, Fujita K, Valles K, Karki S, et al. 17-Beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 2019;69:1504–19.
Darmawan G, Hamijoyo L, Hasan I. Association between serum uric acid and non-alcoholic fatty liver disease: a meta-analysis. Acta Med Indones. 2017;49:136–47.
Sautin YY, Johnson RJ. Uric acid: the oxidant-antioxidant paradox. Nucleosides Nucleotides Nucleic Acids. 2008;27:608–19.
Ishizuka Y, Nakayama K, Ogawa A, Makishima S, Boonvisut S, Hirao A, et al. TRIB1 downregulates hepatic lipogenesis and glycogenesis via multiple molecular interactions. J Mol Endocrinol. 2014;52:145–58.
Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607.
Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–8.
Kraja AT, Borecki IB, Tsai MY, Ordovas JM, Hopkins PN, Lai CQ, et al. Genetic analysis of 16 NMR-lipoprotein fractions in humans, the GOLDN study. Lipids. 2013;48:155–65.
Tilg H, Diehl AM. Cytokines in alcoholic and nonalcoholic steatohepatitis. N Engl J Med. 2000;343:1467–76.
Moschen AR, Molnar C, Enrich B, Geiger S, Ebenbichler CF, Tilg H. Adipose and liver expression of interleukin (IL)-1 family members in morbid obesity and effects of weight loss. Mol Med. 2011;17:840–5.
Chackelevicius CM, Gambaro SE, Tiribelli C, Rosso N. Th17 involvement in nonalcoholic fatty liver disease progression to non-alcoholic steatohepatitis. World J Gastroenterol. 2016;22:9096–103.
Harley IT, Stankiewicz TE, Giles DA, Softic S, Flick LM, Cappelletti M, et al. IL-17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology. 2014;59:1830–9.
Giles DA, Moreno-Fernandez ME, Stankiewicz TE, Cappelletti M, Huppert SS, Iwakura Y, et al. Regulation of inflammation by IL-17A and IL-17F modulates non-alcoholic fatty liver disease pathogenesis. PLoS One. 2016;11:e0149783.
Giles DA, Moreno-Fernandez ME, Stankiewicz TE, Graspeuntner S, Cappelletti M, Wu D, et al. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat Med. 2017;23:829–38.
Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–23.
Genetics Consortium UKIBD, Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–4.
Mukai T, Egawa M, Takeuchi T, Yamashita H, Kusudo T. Silencing of FABP1 ameliorates hepatic steatosis, inflammation, and oxidative stress in mice with nonalcoholic fatty liver disease. FEBS Open Bio. 2017;7:1009–16.
Özenirler S, Degertekin CK, Erkan G, Elbeğ Ş, Tuncer C, Kandilc U, et al. Serum liver fatty acid binding protein shows good correlation with liver histology in NASH. Hepatogastroenterology. 2013;60:1095–100.
Li Q, Qu HQ, Rentfro AR, Grove ML, Mirza S, Lu Y, et al. PNPLA3 polymorphisms and liver aminotransferase levels in a Mexican American population. Clin Invest Med. 2012;35:E237–45.
Battelli MG, Bortolotti M, Polito L, Bolognesi A. The role of xanthine oxidoreductase and uric acid in metabolic syndrome. Biochim Biophys Acta Mol basis Dis. 2018;1864:2557–65.
Gandhy SU, Imanirad P, ** UH, Nair V, Hedrick E, Cheng Y, et al. Specificity protein (Sp) transcription factors and metformin regulate expression of the long non-coding RNA HULC. Oncotarget. 2015;6:26359–72.
Yilmaz Y, Eren F, Colak Y, Senates E, Celikel CA, Imeryuz N. Hepatic expression and serum levels of syndecan 1 (CD138) in patients with nonalcoholic fatty liver disease. Scand J Gastroenterol. 2012;47:1488–93.
Fang L, Zhang M, Li Y, Liu Y, Cui Q, Wang N. PPARgene: a database of experimentally verified and computationally predicted PPAR target genes. PPAR Res. 2016;2016:6042162.
Acknowledgements
“Not applicable”.
Funding
In eMERGE Network (Phase 3 ascertainment), this phase of the eMERGE Network was initiated and funded by the NHGRI through the following grants: U01HG008666 (Cincinnati Children’s Hospital Medical Center); U01HG008657 (Kaiser Washington/University of Washington); U01HG008685 (Brigham and Women’ s Hospital); U01H00G8672 (Vanderbilt University Medical Center); U01HG006379 (Mayo Clinic); U01HG008679 (Geisinger Clinic); U01HG008680 (Columbia University Health Sciences); U01HG008684 (Children’s Hospital of Philadelphia); U01HG008673 (Northwestern University); U01HG008701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG00676 (Partners Health-care/Broad Institute); and U01HG008664 (Baylor College of Medicine). In eMERGE Network (Phase 1 and 2 ascertainment), the eMERGE Network was initiated and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children s Hospital Medical Center/Boston Children’s Hospital); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health (now Kaiser Permanente Washington Health Research Institute)/University of Washington; U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center) with U01HG004438 (CIDR) and U01HG004424 (the Broad Institute) serving as Genoty** Centers. This project was also supported by NIH R01DK099222 (SD), and U01AI130830, P30AR070549, R01AI024717, the US Department of Veterans Affairs (I01BX001834) (JBH).
Author information
Authors and Affiliations
Consortia
Contributions
All authors agree to be accountable for all aspects of the work and read and approved the final manuscript. BN, JBH, and SAX contributed to the study concept and design, data review and comments, interpretation of data, study supervision, and manuscript preparation. BN, TL YH, SP, IBS, DRC, XN, WW, and BB contributed to the data acquisition, data preparation, and data analyses. BN, JJC, FDM, RJC, JAP, ITWH, SD, DSC, EBL, DJC, SV, MDR, AGG, SM, MSW, DRC, GPJ, IJK, HH, and RL contributed to the organization, data preparation, and manuscript review and critique.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Protocols for this study were approved by the Institutional Review Boards (IRBs) at the institutions where participants were recruited; all included participants provided written informed consent prior to inclusion in the study.
Consent for publication
“Not applicable”.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
One excel file with 6 master tables divided into 18 table-sheets. (XLSX 227 kb)
Additional file 2:
Additional methodology. (DOC 53 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Namjou, B., Lingren, T., Huang, Y. et al. GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network. BMC Med 17, 135 (2019). https://doi.org/10.1186/s12916-019-1364-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12916-019-1364-z