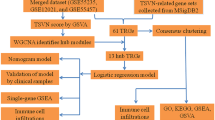
Abstract
Background
Rheumatoid arthritis (RA) and osteoarthritis (OA) share some similar arthritic symptoms, but different mechanisms underlie the pathogenesis of these two diseases. Analysis of differentially expressed molecules in rheumatoid arthritis and osteoarthritis may assist in improving diagnosis and treatment strategies in clinical practice.
Methods
Microarray and RNA-seq data were acquired from the gene expression omnibus database. Differentially expressed genes (DEGs) were identified using Bioconductor packages. Receiver operating characteristic curves were plotted to assess performance. Gene ontology enrichment analysis was conducted using the clusterProfiler application. During validation, synovial fluid was harvested from patients who had undergone in-hospital joint replacement, in which the expression of proteins was measured using enzyme-linked immunosorbent assays.
Results
Compared with OA samples, RA samples showed 14 genes to be upregulated and 3 to be downregulated. Gene ontology analysis indicated that DEGs principally included molecules responsible for the regulation of a synovial tissue inflammatory response. Seven genes displayed a good discriminatory power with an AUC higher than 0.90. ADAMDEC1 was the biomarker that most clearly discriminated RA from OA in the database, exhibiting an AUC of 0.999, a sensitivity of 100%, and a specificity of 97.8%. Following validation, the expression levels of ADAMDEC1 in the synovial fluid from RA patients were significantly higher than those in the synovial fluid from OA patients (P < 0.05). At the cut-off value of 1957 pg/mL, ADAMDEC1 expression in the synovial fluid discriminated RA from OA with an AUC of 0.951, a specificity of 88.6%, and a sensitivity of 92.9%.
Conclusion
The differential expression of genes in RA compared with OA indicates potential targets for molecular diagnosis and treatment. The presence of ADAMDEC1 in synovial fluid is a good biomarker of RA.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a common systemic autoimmune disease. It is characterized by chronic synovial inflammation and hyperplasia in addition to joint destruction, ultimately leading to disability. RA currently affects 0.1%–0.5% of adults [1]. It is approximately three times more common in women than in men, increasing in severity with age, but with a higher incidence in women aged more than 65 years [1, 2]. RA joints are characterized by inflammation of the synovium, which leads to destruction of the articular cartilage and underlying bone when the disease progresses without efficient control. Synovial hyperplasia results from synovial outgrowth or villi which consist of macrophages, cells of the synovial lining, lymphocytes, and blood vessels [3].
Osteoarthritis (OA) is a common form of chronic arthritis in the aged population [4]. There are multiple risk factors for the development of OA, including aging, obesity, and genetic factors [5]. OA is a degenerative joint condition, primarily affecting the hands, hips, and knees in which there is a loss of articular cartilage, synovial membrane dysfunction, sclerosis of the subchondral bone, and osteophyte formation combined with depletion of matrix proteins driven by proteases [15].
Gene ontology (GO) enrichment analyses of DEGs
Annotations of the cellular components (CCs), biological processes (BPs), and molecular functions (MFs) of the DEGs were analyzed using cluster Profiler package in Bioconductor to perform GO enrichment analysis (https://git.bioconductor.org/packages/clusterProfiler).
Receiver Operating Characteristic (ROC) analysis
ROC curves were plotted using the “pROC” package in R software [16]. Optimal sensitivity and specificity were calculated depending on the different cut-off values of gene expression, allowing the discrimination between RA and OA. ROC curves were interpreted in terms of sensitivity, specificity, and area under the ROC curve (AUC).
Synovial fluid collection and measurement of ADAMDEC1 protein
Synovial fluid samples were collected from patients who had undergone joint replacement surgery from May 2016 to December 2019. The diagnosis of OA and RA was based on laboratory examinations, radiological examinations, and described symptoms.
The protein (ADAMDEC1) encoded by the DEG with the most discrimination power was validated in the synovial fluid. After dilution, the expression levels of ADAMDEC1 were measured using an enzyme-linked immunosorbent assay kit (Cusabio, China), in accordance with the manufacturer’s instructions.
To investigate if the secretion of ADAMDEC1 into the synovial fluid was a biomarker to discriminate RA from OA, the AUC, and the sensitivity and specificity, were calculated as described above.
Statistical analysis
The DEGs in the datasets investigated were analyzed using the limma, edgeR, and DESeq2 packages in R software. The significance of differences in ADAMDEC1 protein level in synovial fluid between RA and OA patients was determined via t-test using SPSS v26 (IBM, USA). P-values < 0.05 were considered statistically significant.
Results
Identification of DEGs between RA and OA in tissue from datasets
In the present study, a total of 311 samples of synovial tissue were obtained from datasets, of which 87 samples were from OA patients and 224 were from RA patients (supplementary table 1). Differential expression analysis between OA and RA synovial tissue samples was conducted using the seven datasets obtained from GEO (Fig. 1a). A total of 17 DEGs were identified that displayed significantly different expressions in RA compared with OA samples. In brief, 14 genes were upregulated and 3 were downregulated (Fig. 1b) in RA samples compared with OA samples.
Identification of 17 DEGs between RA and OA. a Histogram indicating the number of overlap** DEGs expressed between RA and OA tissues. The bar chart above shows the DEGs between RA and OA in each type of intersection. The dotted line at the bottom right shows the types of events included in each type of intersection. 17 DEGs were identified in seven transcription profile datasets (GSE55235, GSE55457, GSE55584, GSE1919, GSE36700, GSE12021, and GSE89408) using package UpSetR. b A chart of 17 genes expressed differentially between RA and OA from all datasets (P < 0.05)
Discrimination power of DEGs based on the ROC curve
The discrimination power of the significantly expressed genes was determined based on the AUC, specificity, and sensitivity (Table 1). Every gene of the 17 DEGs could distinguish RA from OA, with an AUC higher than 0.8. Of these, seven DEGs (CXCL13, GPR1, CCL18, ADAMDEC1, GZMB, IL-7R, and RASGRP1) had an AUC higher than 0.90, with a specificity higher than 85%. In particular, ADAMDEC1 was the most powerful gene from the database for the discrimination of RA from OA (AUC: 0.999; sensitivity: 100%; specificity: 97.83%).
GO enrichment analysis of the DEGs
GO analysis demonstrated that the DEGs were principally enriched in BP, MF, and CC terms (Table 2). The BP terms involved in the 17 DEGs were primarily as follows: “T cell differentiation, cell mediated immunity” “inflammatory response” and “response to stimuli” The roles of the 17 DEGs in MF involved activation of “chemokine activity” promotion of “chemokine receptor binding” and regulation of “cytokine activity, receptor binding” and “G protein-coupled receptor binding” The CC terms of the 17 DEGs were related to components primarily located on the “external side of plasma membrane” “clathrin-coated vesicle membrane” and “T cell receptor complex”
Determination of ADAMDEC1 expression in the synovial fluid
Considering that ADAMDEC1 exhibited the greatest power of discrimination in the data analysis, it was predicted that the encoded protein would be expressed in its secretory form and, therefore, be detected in the synovial fluid. Thus, 44 synovial fluid samples from patients were collected in the present study, of which 14 samples were from patients with RA (RA group) and 30 were from patients with OA disease (OA group) (Table 3). Compared with the synovial fluid from the OA group, that from the RA group showed a significantly higher expression level of ADAMDEC (Fig. 2a). With the adoption of a cut-off value of 1957 pg/mL, ADAMDEC1 in the synovial fluid was able to discriminate RA patients from OA patients with an AUC value of 0.951, a specificity of 88.6%, and a sensitivity of 92.9% (Fig. 2b).
Discussion
Although new potential therapeutic targets have been discovered with clinical significance in RA disease progression control, the treatment for OA and RA mainly relieves the symptoms when diseases progress into the late phase. The present study identified 17 DEGs between OA and RA samples in the database, among which one gene product was confirmed to be differently expressed between the RA and OA synovial fluid. The DEGs demonstrated considerable potential for the discrimination of RA from OA, and may be involved in disease pathogenesis; thus, this gene product may act as a new therapeutic target of RA in future.
Gene transcription and expression studies are now widely used to improve diagnosis and confirm novel pathways implicated in the pathogenesis of OA and RA [17, 18]. In previous work by other researchers, a number of DEGs in RA samples were identified in comparison with normal controls, based on limited datasets [19]. In addition, a number of DEGs have been identified in OA samples compared with normal controls [18,19,20,21]. Although it is not difficult for clinicians to distinguish patients with disease status (RA or OA) from healthy individuals, but those suffering from RA have symptoms similar to those with OA, and this results in difficulties in defining personalized treatment strategies for each disease group. While some work has been conducted to identify DEGs between OA tissues and RA tissues [6, 17, 22, 23], researchers have found large numbers of DEGs in small datasets in their studies. In the present study, a total of 17 DEGs were identified from seven datasets, reducing the number of candidate biomarkers for discriminating between OA and RA.
A number of DEGs in the present study, for instance, CXCL13, CD247, GZMB, CCL18, IL7R, and ADAMDEC1 were identical to those reported by Li et al., indicating their conservative role in RA pathogenesis [22]. However, no validation of clinical samples has been previously reported. In the present study, ADAMDEC1 displayed the greatest discrimination between RA and OA in dataset analysis, and the transcription product was detected in the synovial fluid. In addition, the secretion of ADAMDEC1 in the synovial fluid also differed between RA and OA with good sensitivity and specificity. The sampling of synovial fluid is less invasive than that of synovial tissue for patients, indicating that ADAMDEC1 in the synovial fluid could represent a good biomarker for the discrimination of RA from OA.
In the present study, the role of ADAMDEC1 in identifying the presence of RA was indicated by dataset analysis of synovial tissue, eventually confirmed by protein expression in the synovial fluid. As a metalloprotease, ADAMDEC1 is not only associated with a number of inflammatory diseases (including pulmonary sarcoidosis, atherosclerosis, and Crohn’s disease) but it also plays a key role in the pathogenesis of cancer [24,25,26]. In brief, ADAMDEC1 promotes the proliferation, migration, and invasion of glioma, which is mediated by the downregulation of active caspase 3 and active caspase 9 [27]. In cancer stem cells, ADAMDEC1 solubilizes FGF2 to induce FGFR1, upregulating ADAMDEC1 expression to maintain its stemness [28]. Although the role of ADAMDEC1 has not been investigated in RA, its ability to induce inflammation in RA may be achieved by modulating the polarization of M1 macrophages as described in rosacea [29]. Evidence for the high expression of ADAMDEC1 in the synovial fluid has suggested its association with RA; thus, an investigation of the mechanism in the future might establish that it represents a potential target for RA treatment.
Although ADAMEDC1 was not involved in GO enrichment, other DEGs might play key roles in the regulation of RA pathogenesis. In the present study, several significantly expressed genes were found to be involved in immune response, chemokine‐mediated signaling pathway, and inflammatory response, indicating that RA is characterized by autoimmune and inflammatory processes similar to those studied previously [19]. Chemokines, cytokines, and their receptors play a fundamental role in the activation of monocytes and lymphocytes at the site of inflammation, especially in RA [23, 30]. A vicious cycle of changing chemokine levels and signal transduction pathways contributes to cartilage and bone destruction by RA synoviocytes [22, 31]. For example, CCL18 released from synovial macrophages and endothelial cells in RA synovial tissues may activate fibroblast-like synoviocytes and is partly responsible for the pathogenesis of RA [32].
There are also a number of limitations to the present study. First, the number of cases with which validation was conducted was small and from a single center, but the results actually confirm the differential expression of ADAMDEC1 in clinical samples. Second, the RA group studied here was actually derived from patients who received joint replacement, indicating that the progress of disease had not been efficiently controlled. In fact, our findings indicate that elevated ADAMDEC1 in the synovial fluid suggests the presence of RA lesions. The discriminative power for detecting RA lesion in early stage is necessary to be determined in future.
Conclusions
In conclusion, bioinformatics analysis provided 17 potential markers in the synovial tissue for discriminating RA from OA with good sensitivity and specificity. After validation in clinical synovial fluid, ADAMDEC1 was found to be powerful enough to detect RA lesions even in patients who received joint replacement, indicating that it has a potential role as a biomarker for diagnosis and prognosis in the future.
Availability of data and materials
All data generated or analyzed during the study are included in this published article and its supplementary information files.
Abbreviations
- RA:
-
Rheumatoid arthritis
- OA:
-
Osteoarthritis
- DEG:
-
Differentially expressed genes
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the ROC curve
- GO:
-
Gene Ontology
- ADAMDEC1:
-
ADAM Like Decysin 1
- CD247:
-
T-Cell Surface Glycoprotein CD3 Zeta Chain
- CD3D:
-
T‑cell surface glycoprotein CD3 δ chain
- CXCL10:
-
C-X-C Motif Chemokine Ligand 10
- CXCL13:
-
C-X-C Motif Chemokine Ligand 13
- GCH1:
-
GTP Cyclohydrolase 1
- GPR1:
-
G Protein-Coupled Receptor 1
- GZMB:
-
Granzyme B
- GZMH:
-
Granzyme H
- IL7R:
-
Interleukin 7 receptor
- QPCT:
-
Glutaminyl-Peptide Cyclotransferase
- RASGRP1:
-
RAS Guanyl Releasing Protein 1
- SCRG1:
-
Stimulator Of Chondrogenesis 1
- SEL1L3:
-
SEL1L Family Member 3
- TNFAIP3:
-
TNF Alpha Induced Protein 3
- ZIC1:
-
Zic Family Member 1
References
Sparks JA. Rheumatoid Arthritis. Ann Intern Med. 2019;170:Itc1-itc16.
Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388:2023–38.
Beasley J. Osteoarthritis and rheumatoid arthritis: conservative therapeutic management. J Hand Ther. 2012;25:163–71 quiz 72.
McDougall C, Hurd K, Barnabe C. Systematic review of rheumatic disease epidemiology in the indigenous populations of Canada, the United States, Australia, and New Zealand. Semin Arthritis Rheum. 2017;46:675–86.
Bijlsma JW, Berenbaum F, Lafeber FP. Osteoarthritis: an update with relevance for clinical practice. Lancet. 2011;377:2115–26.
Xue F, Zhang C, He Z, Ding L, **ao H. Analysis of critical molecules and signaling pathways in osteoarthritis and rheumatoid arthritis. Mol Med Rep. 2013;7:603–7.
Berenbaum F. New horizons and perspectives in the treatment of osteoarthritis. Arthritis Res Ther. 2008;10(Suppl 2):S1.
Geurts J, Vermeij EA, Pohlers D, Arntz OJ, Kinne RW, van den Berg WB, et al. A novel Saa3-promoter reporter distinguishes inflammatory subtypes in experimental arthritis and human synovial fibroblasts. Ann Rheum Dis. 2011;70:1311–9.
Huber R, Hummert C, Gausmann U, Pohlers D, Koczan D, Guthke R, et al. Identification of intra-group, inter-individual, and gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther. 2008;10:R98.
Lauwerys BR, Hernández-Lobato D, Gramme P, Ducreux J, Dessy A, Focant I, et al. Heterogeneity of synovial molecular patterns in patients with arthritis. PLoS One. 2015;10:e0122104.
Ungethuem U, Haeupl T, Witt H, Koczan D, Krenn V, Huber H, et al. Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis. Physiol Genomics. 2010;42a:267–82.
Woetzel D, Huber R, Kupfer P, Pohlers D, Pfaff M, Driesch D, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther. 2014;16:R84.
Walsh AM, Wechalekar MD, Guo Y, Yin X, Weedon H, Proudman SM, et al. Triple DMARD treatment in early rheumatoid arthritis modulates synovial T cell activation and plasmablast/plasma cell differentiation pathways. PLoS One. 2017;12:e0183928.
Tremblay J, Déziel E. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics. 2010;11:587.
Jia K, Wu Y, Huang J, Wu H. Survival-Associated Alternative Splicing Events in Pan-Renal Cell Carcinoma. Front Oncol. 2019;9:1317.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77.
Lu QY, Han QH, Li X, Li ZC, Pan YT, Liu L, et al. Analysis of differentially expressed genes between rheumatoid arthritis and osteoarthritis based on the gene co-expression network. Mol Med Rep. 2014;10:119–24.
Xu Y, Huang Y, Cai D, Liu J, Cao X. Analysis of differences in the molecular mechanism of rheumatoid arthritis and osteoarthritis based on integration of gene expression profiles. Immunol Lett. 2015;168:246–53.
**ong Y, Mi BB, Liu MF, Xue H, Wu QP, Liu GH. Bioinformatics Analysis and Identification of Genes and Molecular Pathways Involved in Synovial Inflammation in Rheumatoid Arthritis. Med Sci Monit. 2019;25:2246–56.
Cai P, Jiang T, Li B, Qin X, Lu Z, Le Y, et al. Comparison of rheumatoid arthritis (RA) and osteoarthritis (OA) based on microarray profiles of human joint fibroblast-like synoviocytes. Cell Biochem Funct. 2019;37:31–41.
Lin J, Wu G, Zhao Z, Huang Y, Chen J, Fu C, et al. Bioinformatics analysis to identify key genes and pathways influencing synovial inflammation in osteoarthritis. Mol Med Rep. 2018;18:5594–602.
Li WC, Bai L, Xu Y, Chen H, Ma R, Hou WB, et al. Identification of differentially expressed genes in synovial tissue of rheumatoid arthritis and osteoarthritis in patients. J Cell Biochem. 2019;120:4533–44.
Zhang R, Yang X, Wang J, Han L, Yang A, Zhang J, et al. Identification of potential biomarkers for differential diagnosis between rheumatoid arthritis and osteoarthritis via integrative genome-wide gene expression profiling analysis. Mol Med Rep. 2019;19:30–40.
O’Shea NR, Chew TS, Dunne J, Marnane R, Nedjat-Shokouhi B, Smith PJ, et al. Critical Role of the Disintegrin Metalloprotease ADAM-like Decysin-1 [ADAMDEC1] for Intestinal Immunity and Inflammation. J Crohns Colitis. 2016;10:1417–27.
Sugimoto N, Nakayama T, Kasai Y, Asaka D, Mitsuyoshi R, Tsurumoto T, et al. The role of ADAM-like decysin 1 in non-eosinophilic chronic rhinosinusitis with nasal polyps. Acta Otolaryngol. 2018;138:830–6.
Crouser ED, Culver DA, Knox KS, Julian MW, Shao G, Abraham S, et al. Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am J Respir Crit Care Med. 2009;179:929–38.
Liu X, Huang H, Li X, Zheng X, Zhou C, Xue B, et al. Knockdown of ADAMDEC1 inhibits the progression of glioma in vitro. Histol Histopathol. 2020;35:997–1005.
Jimenez-Pascual A, Hale JS, Kordowski A, Pugh J, Silver DJ, Bayik D, et al. ADAMDEC1 Maintains a Growth Factor Signaling Loop in Cancer Stem Cells. Cancer Discov. 2019;9:1574–89.
Liu T, Deng Z, **e H, Chen M, Xu S, Peng Q, et al. ADAMDEC1 promotes skin inflammation in rosacea via modulating the polarization of M1 macrophages. Biochem Biophys Res Commun. 2020;521:64–71.
Choi J, Selmi C, Leung PSC, Kenny TP, Roskams T, Gershwin ME. Chemokine and chemokine receptors in autoimmunity: the case of primary biliary cholangitis. Expert Rev Clin Immunol. 2016;12:661–72.
Dayer JM. The saga of the discovery of IL-1 and TNF and their specific inhibitors in the pathogenesis and treatment of rheumatoid arthritis. Joint Bone Spine. 2002;69:123–32.
Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol. 2005;78:14–26.
Acknowledgements
Not applicable.
Funding
This study was supported by Science and Technology Bureau of **hua (2021–4-135).
Author information
Authors and Affiliations
Contributions
KL collected and analyzed the data from the database. WL and DC collected the synovial fluid from patients and participated in data analysis. PX was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was conducted in accordance with the relevant guidelines and regulations. Verbal consent was obtained from all participants and this form of consent acquisition was approved by the Ethics Committee of the Affiliated Dongyang Hospital of Wenzhou Medical University (2016-YB002). All experimental protocols were approved by Ethics Committee of the Affiliated Dongyang Hospital of Wenzhou Medical University (2016-YB002).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplemental Table 1.
Number of samples in downloadeddataset.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kang, L., Dai, C., Wang, L. et al. Potential biomarkers that discriminate rheumatoid arthritis and osteoarthritis based on the analysis and validation of datasets. BMC Musculoskelet Disord 23, 319 (2022). https://doi.org/10.1186/s12891-022-05277-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12891-022-05277-x