Abstract
Long noncoding RNAs (lncRNAs) are endogenous RNA transcripts longer than 200 nucleotides which regulate epigenetically the expression of genes but do not have protein-coding potential. They are emerging as potential key regulators of diabetes mellitus and a variety of cardiovascular diseases. Diabetic cardiomyopathy (DCM) refers to diabetes mellitus-elicited structural and functional abnormalities of the myocardium, beyond that caused by ischemia or hypertension. The purpose of this review was to summarize current status of lncRNA research for DCM and discuss the challenges and possible strategies of lncRNA research for DCM. A systemic search was performed using PubMed and Google Scholar databases. Major conference proceedings of diabetes mellitus and cardiovascular disease occurring between January, 2014 to August, 2018 were also searched to identify unpublished studies that may be potentially eligible. The pathogenesis of DCM involves elevated oxidative stress, myocardial inflammation, apoptosis, and autophagy due to metabolic disturbances. Thousands of lncRNAs are aberrantly regulated in DCM. Manipulating the expression of specific lncRNAs, such as H19, metastasis-associated lung adenocarcinoma transcript 1, and myocardial infarction-associated transcript, with genetic approaches regulates potently oxidative stress, myocardial inflammation, apoptosis, and autophagy and ameliorates DCM in experimental animals. The detail data regarding the regulation and function of individual lncRNAs in DCM are limited. However, lncRNAs have been considered as potential diagnostic and therapeutic targets for DCM. Overexpression of protective lncRNAs and knockdown of detrimental lncRNAs in the heart are crucial for defining the role and function of lncRNAs of interest in DCM, however, they are technically challenging due to the length, short life, and location of lncRNAs. Gene delivery vectors can provide exogenous sources of cardioprotective lncRNAs to ameliorate DCM, and CRISPR–Cas9 genome editing technology may be used to knockdown specific lncRNAs in DCM. In summary, current data indicate that LncRNAs are a vital regulator of DCM and act as the promising diagnostic and therapeutic targets for DCM.
Similar content being viewed by others
Background
Diabetic cardiomyopathy (DCM) refers to diabetes-associated changes in the structure and function of the myocardium that are not directly attributable to other confounding factors such as coronary heart disease or hypertension [1]. It is estimated that DCM occurs in approximately 12% of diabetic patients [2]. DCM is associated with the development of overt heart failure and worse prognosis of diabetic patients [3, 4]. A strategy for prevention and treatment in order to improve the prognosis of DCM has not been established [5,6,7].
Long noncoding RNAs (lncRNAs) are RNA transcripts longer than 200 nucleotides which, although not having the function of direct coding proteins, can regulate the expression of genes at transcriptional, post-transcriptional, and translational levels [8]. Over the past decade, lncRNAs have received widespread attention as potentially new and crucial players of biological regulation [9, 10]. Their cell-type and tissue-specific expression in health and cardiovascular disease provides the avenue for the diagnosis and treatment of cardiovascular disease [11, 12]. Emerging studies find that lncRNAs are aberrantly regulated in DCM, and impacting the expression of specific lncRNAs is capable of regulating the pathophysiological process of DCM [13,14,15]. Although the detailed data regarding the role of specific lncRNAs in DCM are limited, they are increasingly identified as a vital regulator of DCM in experimental animals. To get insight into current status of lncRNA research for DCM, we used PubMed and Google Scholar databases to search systemically the published articles that are involved in lncRNAs and DCM. Major conference proceedings of diabetes mellitus and cardiovascular disease occurring between January, 2014 to August, 2018 were also searched to identify unpublished studies that may be potentially eligible. Based on the data obtained from these databases, we present an overview of lncRNA research for DCM. We also discuss the challenges and possible strategies of lncRNAs as diagnostic and therapeutic targets for DCM.
Diabetes-induced cardiac damage
Diabetes mellitus affects the heart through various mechanisms including metabolic disturbance (suppressed glucose oxidation, enhanced fatty acid metabolism, hyperinsulinemia, insulin resistance, accumulation of advanced glycation end-products, etc.), subcellular component abnormalities, microvascular impairment, and autonomic dysfunction [16, 17]. Eventually myocardium develops local inflammation, coronary arterial endothelial dysfunction, necrosis, apoptosis, autophagy, fibrosis, atherosclerosis, steatosis, and ventricular hypertrophy (Fig. 1) [18, 19]. These pathological changes in the structure, morphology, and function of the heart develop in diabetic patients, especially patients with type 2 diabetes mellitus (T2DM), even without the presence of ischemic heart disease and hypertension, termed diabetic cardiomyopathy (DCM) [1]. It is estimated that DCM occurs in approximately 12% of diabetic patients [2]. Clinical studies indicate that DCM increases the risk of overt heart failure and worsens the prognosis in diabetic patients [3, 4].
Pathogenesis of diabetic cardiomyopathy. In diabetes mellitus, repressed glucose oxidation, enhanced fatty acid metabolism, hyperinsulinemia, insulin resistance, and accumulation of advanced glycation end-products lead to oxidative stress, microcirculation impairment, mitochondrial dysfunction, and autonomic neuropathy. These pathogenic factors together result in myocardial inflammation, endothelial dysfunction, necrosis, apoptosis, autophagy, fibrosis, athrosclerosis, and cardiac hypertrophy, impair Ca2+ homeostasis, and activate the renin-angiotensin system (RAS). Eventually these pathogenic changes in the myocardium impair the diastolic and systolic function of the heart
Animal models of DCM are critically important for us to advance the understanding of pathogenic mechanisms of DCM and discover new diagnostic and therapeutic targets for DCM. Over the past thirty years, investigators have developed many rodent models of diabetes mellitus and DCM [19, 20]. They are able to provide many advantages in the availability of adequate healthy controls and the absence of confounding factors such as marked differences in age, concomitant pathologies, and pharmacological treatments. Among these models, streptozotocin (STZ)-induced cardiomyopathy of type 1 diabetes mellitus (T1DM) and leptin receptor deficient (db/db)- or leptin deficient (ob/ob)-cardiomyopathy of T2DM are frequently used in the study of lncRNAs [5, 20, 21].
LncRNAs in the heart
LncRNAs represent one of the most prominent but least understood transcriptome in the heart. According to the NONCODE database (http:www.noncode.org/, version 5), there are 172,216 and 131,697 lncRNA transcripts for humans and mice, respectively. Thousands of lncRNAs have been identified to express abundantly in the myocardial tissues [22,23,24]. Many of these lncRNAs are dynamically transcribed during the development, differentiation, and maturation of cardiac myocytes [25,26,27].
LncRNAs have been known to control and regulate the expression of broad ranges of genes in cardiomyocytes [28, 29]. Similar to protein-coding RNAs, individual lncRNAs have specific subcellular distribution that is critical for their functions [30, 31]. Some lncRNAs are enriched in the nucleus and are involved in regulating nuclear processes, such as DNA replication-associated biological processes, mRNA transcription, and RNA processing [23, 32]. In the nucleus, lncRNAs can interact with DNA to form RNA-DNA complexes to reprogram gene expression, act as molecular scaffold, activate or suppress transcription [33, 34]. Other lncRNAs are enriched in the cytoplasm where they can impact protein localization or modulate mRNA stability and translation [35]. LncRNAs can also bind mRNA transcripts to either stabilize or promote translation, cause steric hindrance to block translation (e.g., acting as decoys), regulate RNA splicing and stability, and act as a sponge for microRNAs [36,37,38]. In the cytoplasm, lncRNAs can interact with proteins to mediate protein trafficking and signaling and impact the function of bound proteins [39].
LncRNA-mediated regulation of gene expression in the heart has been known to involve a variety of mechanisms [40, 41]. Some lncRNAs (for example, cardiac-specific lncRNA Myheart) can interact with chromatin remodeling factors to reprogram gene expression [28]. Some lncRNAs (e.g., the lncRNA Braveheart) can guide chromatin-modifying complexes to their required genomic destination and serve as docking stations for complex recruitment (acting as scaffolding) [42]. Certain lncRNAs (e.g., the cardiac-enriched lncRNA Upperhad) activate transcription of certain genes by guiding transcription factors to their promoters [43, 44]. Particular lncRNAs (e.g., the lncRNA cardiac autophagy inhibitory factor) are capable of suppressing transcription by sequestering transcription factors [45]. Some lncRNAs (e.g., the lncRNA myocardial infarction-associated transcript [MIAT]) can bind to complementary microRNAs (e.g., microRNA-24) via base pairing to sequester them (acting as “microRNA sponges”) [46]. Various lncRNAs (e.g., the lncRNA metastasis-associated lung adenocarcinoma transcript 1 [MALAT1]) can interact with mRNA to regulate their translation and splicing [47, 48]. Other lncRNAs (e.g., cardiac autophagy inhibitory factor) can interact with proteins to mediate their trafficking and signaling and regulate the function of bound proteins [45].
LncRNAs play crucial roles in various cardiac diseases [38, 45, 49, 50]. LncRNAs can be targeted to change the physiological function of cardiac myocytes [51, 52]. In cardiac disease, lncRNAs are regulated in a cell type/tissue-specific manner [53, 54]. Manipulating the expression of specific lncRNAs with genetic and pharmacological approaches impacts the severity of myocardial ischemia/reperfusion injury, cardiac hypertrophy, heart failure, and diabetic vascular complications. Thus, certain lncRNAs that are conserved in the heart may have therapeutic potential on various heart diseases [12, 14, 55]. Moreover, some circulating lncRNAs have been proposed to be the biomarker of cardiac disease [56].
Regulation and function of specific lncRNAs in DCM
Specific lncRNAs have been identified to express differentially in the heart with DCM [15, 37, 57, 58]. The aberrant expression of specific lncRNAs is associated with the pathophysiological process of DCM, such as oxidative stress, inflammation, apoptosis, myocardial fibrosis, and autophagy (Fig. 2) [15, 37, 57, 58]. Manipulating specific lncRNAs to alter their expression is able to ameliorate DCM [37, 57, 58]. Despite the limited data regarding the regulation and function of specific lncRNAs in DCM, lncRNAs are considered as a promising target/candidate for the treatment and diagnosis of DCM. In this section, we discuss several of the lncRNAs that may have a good potential as a target/candidate for the treatment and diagnosis of DCM (Table 1).
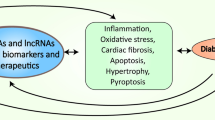
Long noncoding RNAs (lncRNAs) impact the pathophysiological process of diabetic cardiomyopathy. Long noncoding RNAs are regulated in diabetic cardiomyopathy. Changes in the expression of long noncoding RNAs in myocardial tissues influence oxidative stress, myocardial inflammation, cardiomyocyte apoptosis, autophagy, and microvascular impairments. MALAT1: metastasis-associated lung adenocarcinoma transcript 1; MIAT: myocardial infarction-associated transcript; MT-LIPCAR: the mitochondrially encoded long non-coding cardiac associated RNA; SENCR: smooth muscle and endothelial cell-enriched migration/differentiation-associated long noncoding RNA
H19
H19 is a 2.3-kb lncRNA which is transcribed from H19/insulin-like growth factor-II (IGF2) genomic imprinted cluster located on human chromosome 11p15.5 (syntenic to mouse chromosome 7) [59]. H19 and IGF2 genes are expressed in a monoallelic fashion from the maternal and paternal chromosomes, respectively [60, 61]. H19 is transcribed by a polymerase II [62]. H19 transcripts start from the blastocyst stage and reach a high level in the tissues of endodermal, mesodermal, and ectodermal origins [63]. After the birth, H19 expression will be inhibited in most of mammalian tissues [64]. However, H19 remains in high accumulation in mature myocardium of both mice and humans possibly due to enhanced RNA stabilization during cardiomyocyte differentiation [65]. Both primary sequence and secondary structures of H19 show a great extent of conservation among mammals [66].
H19 has recently been identified as an important regulator of the cardiomyopathy of T1DM in experimental rats [57, 58]. Sprague-Dawley rats injected with STZ developed the cardiomyopathy of T1DM with decreased expression of cardiac H19 [57, 58]. Overexpression of H19 in myocardial tissues caused decreases in oxidative stress, inflammation, apoptosis, and autophagy, leading to the amelioration of DCM [57, 58].
H19 serves as template for microRNA-675 expression from H19 first exon [67, 68]. Since microRNA-675 has multiple targets in diverse signaling pathways, H19 is able to regulate a number of biological processes via microRNA-675. For example, the H19/microRNA-675 reduces high glucose-induced apoptosis by targeting voltage-dependent anion channel 1 which is a critical protein required for the mitochondria-mediated apoptosis [58, 69]. In addition, by down-regulating GTP-binding protein Di-Ras-3, the H19/microRNA-675 promotes the phosphorylation of the mechanistic target of rapamycin and inhibits activated autophagy in cardiomyocytes exposed to high glucose [57]. Another pattern of H19 exerting its function is through interacting with proteins and microRNAs. H19 is capable of being folded into a special secondary structure, which allows it to serve as a platform and collect relative proteins [70]. Multiple proteins have been identified to associate with H19, including the RNA binding proteins, KH-type splicing regulatory proteins, inner membrane protease 1, the Hu family of RNA-binding proteins, heterogeneous nuclear ribonucleoprotein U, polypyrimidine tract-binding protein 1, the DNA/chromatin modification factors, S-adenyl-L-homocysteine hydrolase, polycomb repressive complex 2, p53, and isoleucyl tRNA synthetase of mitochondria [68]. These proteins are actively involved in a wide variety of physiological and pathological processes, such as RNA metabolism, gene transcription, and epigenetic modification [68]. MicroRNAs are another group of partners that are essential for H19 to exert its function. It is evident that H19 interacts with Let-7, microRNA-138, microRNA-200a, microRNA-106a, and microRNA-141 [68].
IGF2 proteins are an important growth factor during pregnancy, where they promote both fetal and placental growth [71, 72]. However, the overexpression of IGF2 and its receptors in acute hyperglycemia and diabetes is associated with the progression of DCM by triggering cardiac hypertrophy and apoptosis [73]. The effect of H19 overexpression on the levels of myocardial IGF2 in adults remains unclear. In embryos, the overexpression of H19 results in a decrease in IGF2 expression due to a cis effect of the H19 locus on the adjacent IGF2 gene [74]. It is reasonably believed that IGF2 levels are decreased too in H19-overexpressing animals, and decreased IGF2 contributes to the beneficial effects of H19 overexpression on DCM.
In summary, cardiac H19 is downregulated in DCM, and transgenic overexpression of H19 improves DCM by attenuation of myocardial oxidative stress, inflammation, apoptosis, and autophagy.
MALAT1
MALAT1 is a nuclear transcript localized to the nuclear speckles, a nuclear domain for storage and/or the sites of pre-mRNA splicing [75]. Pre-mRNAs splicing is a pivotal step between transcription and translation of most eukaryotic mRNAs [76]. MALAT1 interacts with several serine/arginine proteins, such as serine/arginine-rich splicing factors and spliceosomal proteins, to regulate pre-mRNA splicing [77,78,79]. In addition, MALAT1 is involved in nuclear organization and epigenetic modulation of gene expression [80, 81]. MALAT1 was abundantly expressed in cardiac myocytes and highly conserved across mammalian species [82, 83]. In the rat cardiomyopathy of T1DM induced by streptozotocin, MALAT1 in myocardial tissues was up-regulated [15, 84]. The knockdown of MALAT1 with the small interfering RNA to attenuate the expression of MALAT1 in diabetic hearts significantly attenuated inflammation and apoptosis and improved DCM [15, 84]. Thus, the upregulation of MALAT1 represents a critical pathogenic mechanism for DCM.
In short, cardiac MALAT1 is upregulated in DCM, and the knockdown of MALAT1 improves DCM by attenuation of myocardial inflammation and apoptosis.
MIAT
MIAT is first identified to be associated with myocardial infarction in a genome-wide association study in 2006 [85]. Before that, MIAT was also known as RNCR2, 2 AK02836 or GOMAFU. MIAT may function as a competing endogenous RNA to upregulate the expression of death-associated protein kinase-2 by sponging miR-22-3p, which consequently leads to the apoptosis of cardiac myocytes [37]. Like MALAT1, the expression of cardiac MIAT was significantly upregulated in Sprague-Dawley rats with the cardiomyopathy of T1DM [37]. The knockdown of MIAT with MIAT-shRNA resulted in improvement of DCM and reduction of apoptosis of cardiac myocytes [37]. The inhibitory effect of MIAT knockdown on apoptosis is attributed to a decrease in the expression of death-associated protein kinase-2. Taken together, the upregulation of cardiac MIAT contributes to the pathogenesis of DCM.
Smooth muscle and endothelial cell-enriched migration/differentiation-associated long noncoding RNA (SENCR)
SENCR is a vascular cell-enriched lncRNA [86]. It promotes the proliferation and migration of smooth muscle cells through regulation of forkhead box protein O1 and transient receptor potential cation channel 6. However, SENCR was down-regulated in T2DM db/db mice and in vascular smooth muscle cells exposed to high glucose [87]. The overexpression of SENCR reversed the inhibitory effect of high glucose on the proliferation and migration of mouse vascular smooth muscle cells. Both clinical and experimental studies indicate that impaired vascular smooth muscle cells by diabetes and high glucose contribute to the increased incidence of DCM [88]. Although there are no reports about the direct impacts of SENCR on DCM, the downregulation of cardiac SENCR may contribute to the pathogenesis of DCM.
The mitochondrially encoded long non-coding cardiac associated RNA (MT-LIPCAR)
MT-LIPCAR (uc022bqs.1, Gene ID: 103504742) is a 781-nucleotide lncRNA which is possibly transcribed from mitochondrial DNA [89]. It can cross the membrane barrier and is released into the circulation. Although there are a large number of RNase in plasma [90], MT-LIPCAR is stable in blood serum/plasma [13, 49, 91]. Recently, de Gonzalo-Calvo et al. analyzed lncRNAs derived from the serum of 48 patients with cardiomyopathy of T2DM and 12 healthy volunteers [13]. MT-LIPCAR levels in plasma were positively associated with left ventricular diastolic dysfunction. Moreover, MT-LIPCAR was strongly correlated with waist circumference, plasma fasting insulin, subcutaneous fat volume, and high-density lipoproteins-C. Collectively, MT-LIPCAR may be an independent predictor of diastolic dysfunction in T2DM patients with DCM [13].
In the clinic, the specific diagnosis of DCM is difficult, since the patients are asymptomatic in the early and middle stages and may concomitantly suffer from ischemic heart disease or hypertension during the late stage [7, 92, 93]. The significant increase in the levels of specific lncRNAs in serum/plasma of patients with DCM, such as MT-LIPCAR, could make lncRNAs specific biomarkers for the diagnosis and prognosis of DCM. A clinical trial recently suggests that MT-LIPCAR in plasma may serve as a promising biomarker of DCM [13]. The value of MT-LIPCAR and other circulating lncRNAs as diagnostic and prognostic markers in DCM needs to be validated. Large multicenter randomized, controlled trials with MT-LIPCAR need to be conducted in patients with DCM.
Antisense non-coding RNA in the INK4 locus (ANRIL)
ANRIL [alias cyclin dependent kinase inhibitor 2B antisense RNA 1 (CDKN2B-AS1) and P15 antisense RNA (P15AS)] is a 3.8 kb lncRNA transcribed from the short arm of human chromosome 9 on p21.3 [94]. ANRIL and the adjacent protein coding genes, cyclin dependent kinase inhibitor 2A (CDKN2A) and cyclin dependent kinase inhibitor 2B (CDKN2B), locate on chromosome 9p21 [95]. The CDKN2A gene encodes several transcripts/proteins, the p16 protein of which functions as inhibitors of cyclin-dependent kinase 4 [96, 97]. The CDKN2B gene encodes cyclin-dependent kinase 4 inhibitor B that functions as a cell growth regulator that control cell cycle G1 progression [98]. ANRIL is an antisense of the CDKN2B gene and is transcribed by RNA polymerase II and spliced into multiple linear and circular isoforms in a tissue-specific manner [99]. ANRIL is capable of recruiting polycomb group proteins to modify the epigenetic chromatin state and binding to a site or sequence to regulate gene expression [100]. It is well known to know that single nucleotide polymorphisms in the human chromosome 9p21 locus are associated with diabetes, cardiovascular disease, and multiple cancers [101,102,103,104,105,106]. Recent studies have identified ANRIL as a highly susceptible region for T2DM, coronary artery disease, and hypertension [107]. Although there is no report regarding the role of ANRIL in DCM, it is reasonably believed that ANRIL might be involved in the pathogenesis of DCM.
In summary, ANRIL is a potential candidate that is associated with the pathogenesis of DCM.
Challenges and potential strategies of lncRNA research for DCM
LncRNAs may be a promising target and/or candidate as biomarkers of DCM diagnosis and for the treatment of DCM. However, at present the function and regulation of thousands of lncRNAs in DCM are still ambiguous. Recently, we performed a systemic microarray-based analysis of the cardiac expression profiles of lncRNAs in T2DM db/db mice on a genetic background of C57BL/6 mice with and without DCM. Among the 23,578 lncRNAs identified, 1479 were differentially expressed in the myocardium of db/db mice between with DCM and without DCM [108]. These results suggest that at least 1479 lncRNAs might be involved in DCM in obese type 2 db/db mice. Determining the individual functionality of these lncRNAs is important for good understanding of cardiac developmental biology and DCM. For the study of individual lncRNAs in DCM, the following questions should be considered: Do lncRNAs contribute to the pathogenesis of DCM? How stable are the lncRNAs in circulation? Is their stability altered in diabetes mellitus and cardiac dysfunction? Are lncRNAs toxic? What are the pharmacokinetics of the lncRNAs? Answering these questions will be important as we study the individual lncRNAs and their role in diagnosis and treatment of DCM.
Some lncRNAs are protective to DCM, such as H19. These lncRNAs are down-regulated in DCM [57], and their overexpression in the heart is considered as a therapeutic strategy for DCM [58]. Owing to the length of lncRNA molecules their overexpression in cardiomyocytes is a complicated matter. Moreover, the long modified transcript is difficult to cross the membrane barrier. Thus, its efficient in vivo delivery would be difficult. Recent studies have reported that gene delivery vectors are capable of provide exogenous expression of the desired lncRNAs [38]. Utilization of gene delivery vectors, like engineered adeno-associated virus, is an alternative approaches to increase the expression of protective lncRNAs in the heart to ameliorate DCM.
Up-regulation of detrimental lncRNAs in DCM, such as MALAT1 and MIAT, could make them promising therapeutics targets for DCM [109]. However, in vivo inhibition of detrimental lncRNAs is a challenge mainly due to their short half live as they are easily degraded by nucleases in bio fluids and the length of lncRNA transcripts. At present, the approaches which are used to manipulate lncRNAs in vivo include mainly the use of small interfering RNAs, antisense oligonucleotides, and the 5′ and 3′ end-modified antisense oligonucleotides, GapmeRs [53, 110]. Each of these approaches have their own advantages and disadvantages. Small interfering RNAs specifically bind to complementary sequences and inhibit the expression of lncRNA targets [111, 112]. Antisense oligonucleotides are capable of targeting specific genes or transcripts directly through Watson-Crick base pairing, and they thus can reduce the levels of lncRNAs of interest [113]. Locked nucleic acid GapmeRs can modulate target lncRNA expression, block lncRNA activity, or induce enzyme-mediated degradation [53, 114]. Despite the potential therapeutic value of small interfering RNAs, antisense oligonucleotides, and GapmeRs in treating human disease, the effects of these approaches may have varied efficacy within the cell due to poor accessibility. Many studies have made use of antisense oligonucleotides to knockdown lncRNAs successfully for functional studies in mice or rats [115,116,117]. Compared with small interfering RNAs, antisense oligonucleotides are able be a better approach since cytoplasmic lncRNAs are efficiently ablated using small interfering RNA. To inhibit upregulated lncRNAs that show co-localization, the hybrid approach works the best [111].
Some lncRNAs are refractory to inhibition by either antisense oligonucleotides or small interfering RNAs. This may be related to the subcellular localization of the lncRNAs, which is not accessible to either RNase H or the interfering RNA machinery [111]. Another cause may be that the lncRNAs are highly structured or blocked due to excessive protein binding or hybridizing to other cellular nucleic acids. To overcome these hurdles, it is necessary to produce a high-throughput method to delete lncRNAs. Emerging studies suggest that CRISPR–Cas9 genome editing technology is able to quickly and effectively delete lncRNAs [118, 119]. Despite no reports about the utilization of CRISPR–Cas9 genome editing technology in DCM, this technology is a potential tool to delete the lncRNAs of interest and modulate the expression of lncRNAs in DCM.
In short, both overexpression of protective lncRNAs and knockdown of detrimental lncRNAs in the heart are crucial for defining the role and function of the lncRNAs of interest in DCM. Either approach is technically challenging due to the length, short life, and location of the lncRNAs of interest. In addition to traditional utilization of small interfering RNAs, antisense oligonucleotides, and GapmeRs to inhibit the lncRNAs of interest, CRISPR–Cas9 genome editing technology is a potential tool to knockdown specific lncRNAs.
Conclusions
LncRNAs play vital roles in the pathogenesis of DCM. Manipulating specific lncRNAs with pharmacological and genetic approaches to alter their expression impacts the development of DCM. In spite of limited data of specific lncRNAs in DCM, they are the potential targets/candidates for DCM. The future research needs to elucidate the regulation, function, and action mechanisms of more lncRNAs in the pathogenesis of DCM to search potential targets/candidates as diagnostic biomarkers of DCM and potential treatment of DCM.
Abbreviations
- ANRIL:
-
Antisense non-coding RNA in the INK4 locus
- DCM:
-
Diabetic cardiomyopathy
- IGF2:
-
Insulin-like growth factor-II
- lncRNAs:
-
Long noncoding RNAs
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript
- MIAT:
-
Myocardial infarction-associated transcript
- MT-LIPCAR:
-
The mitochondrially encoded long non-coding cardiac associated RNA
- SENCR:
-
Smooth muscle and endothelial cell-enriched migration/differentiation-associated long noncoding RNA
- STZ:
-
Streptozotocin
- T1DM:
-
Type 1 diabetes mellitus
- T2DM:
-
Type 2 diabetes mellitus
References
Jia G, Hill MA, Sowers JR. Diabetic cardiomyopathy: an update of mechanisms contributing to this clinical entity. Circ Res. 2018;122:624–38.
Trachanas K, Sideris S, Aggeli C, Poulidakis E, Gatzoulis K, Tousoulis D, Kallikazaros I. Diabetic cardiomyopathy: from pathophysiology to treatment. Hell J Cardiol. 2014;55:411–21.
Qazi MU, Malik S. Diabetes and cardiovascular disease: original insights from the Framingham heart study. Glob Heart. 2013;8:43–8.
Marcinkiewicz A, Ostrowski S, Drzewoski J. Can the onset of heart failure be delayed by treating diabetic cardiomyopathy? Diabetol Metab Syndr. 2017;9:21.
Baumgardt SL, Paterson M, Leucker TM, Fang J, Zhang DX, Bosnjak ZJ, Warltier DC, Kersten JR, Ge ZD. Chronic co-administration of sepiapterin and L-citrulline ameliorates diabetic cardiomyopathy and myocardial ischemia/reperfusion injury in obese type 2 diabetic mice. Circ Heart Fail. 2016;9:e002424.
Gilca GE, Stefanescu G, Badulescu O, Tanase DM, Bararu I, Ciocoiu M. Diabetic cardiomyopathy: current approach and potential diagnostic and therapeutic targets. J Diabetes Res. 2017;2017:1310265.
Lorenzo-Almoros A, Tunon J, Orejas M, Cortes M, Egido J, Lorenzo O. Diagnostic approaches for diabetic cardiomyopathy. Cardiovasc Diabetol. 2017;16:28.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9.
Kataoka M, Wang DZ. Non-coding RNAs including mirnas and lncrnas in cardiovascular biology and disease. Cell. 2014;3:883–98.
Haemmig S, Feinberg MW. Targeting lncRNAs in cardiovascular disease: options and expeditions. Circ Res. 2017;120:620–3.
Sallam T, Sandhu J, Tontonoz P. Long noncoding RNA discovery in cardiovascular disease: decoding form to function. Circ Res. 2018;122:155–66.
de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, Smit JW, Lamb HJ, Llorente-Cortes V, Thum T. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep. 2016;6:37354.
Boon RA, Jae N, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67:1214–26.
Zhang M, Gu H, Chen J, Zhou X. Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int J Cardiol. 2016;202:753–5.
DeFronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ, Hu FB, Kahn CR, Raz I, Shulman GI, Simonson DC, Testa MA, Weiss R. Type 2 diabetes mellitus. Nat Rev Dis Primers. 2015;1:15019.
Lee WS, Kim J. Diabetic cardiomyopathy: where we are and where we are going. Korean J Intern Med. 2017;32:404–21.
Miki T, Yuda S, Kouzu H, Miura T. Diabetic cardiomyopathy: pathophysiology and clinical features. Heart Fail Rev. 2013;18:149–66.
Fuentes-Antras J, Picatoste B, Gomez-Hernandez A, Egido J, Tunon J, Lorenzo O. Updating experimental models of diabetic cardiomyopathy. J Diabetes Res. 2015;2015:656795.
Wu HE, Baumgardt SL, Fang J, Paterson M, Liu Y, Du J, Shi Y, Qiao S, Bosnjak ZJ, Warltier DC, Kersten JR, Ge ZD. Cardiomyocyte GTP cyclohydrolase 1 protects the heart against diabetic cardiomyopathy. Sci Rep. 2016;6:27925.
Ge ZD, Li Y, Qiao S, Bai X, Warltier DC, Kersten JR, Bosnjak ZJ, Liang M. Failure of isoflurane cardiac preconditioning in obese type 2 diabetic mice involves aberrant regulation of microRNA-21, endothelial nitric-oxide synthase, and mitochondrial complex I. Anesthesiology. 2018;128:117–29.
Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, Rodriguez Esteban C, Campistol JM, Yeo GW, Izpisua Belmonte JC. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131:1278–90.
Touma M, Kang X, Zhao Y, Cass AA, Gao F, Biniwale R, Coppola G, **ao X, Reemtsen B, Wang Y. Decoding the long noncoding RNA during cardiac maturation: a roadmap for functional discovery. Circ Cardiovasc Genet. 2016;9:395–407.
Tang Z, Wu Y, Yang Y, Yang YT, Wang Z, Yuan J, Yang Y, Hua C, Fan X, Niu G, Zhang Y, Lu ZJ, Li K. Comprehensive analysis of long non-coding RNAs highlights their spatio-temporal expression patterns and evolutional conservation in sus scrofa. Sci Rep. 2017;7:43166.
He C, Hu H, Wilson KD, Wu H, Feng J, **a S, Churko J, Qu K, Chang HY, Wu JC. Systematic characterization of long noncoding RNAs reveals the contrasting coordination of cis- and trans-molecular regulation in human fetal and adult hearts. Circ Cardiovasc Genet. 2016;9:110–8.
Li Y, Zhang J, Huo C, Ding N, Li J, **ao J, Lin X, Cai B, Zhang Y, Xu J. Dynamic organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine. 2017;24:137–46.
Beermann J, Kirste D, Iwanov K, Lu D, Kleemiss F, Kumarswamy R, Schimmel K, Bar C, Thum T. A large shRNA library approach identifies lncRNA Ntep as an essential regulator of cell proliferation. Cell Death Differ. 2018;25:307–18.
Chang CP, Han P. Epigenetic and lncRNA regulation of cardiac pathophysiology. Biochim Biophys Acta. 2016;1863:1767–71.
Li Y, Du W, Zhao R, Hu J, Li H, Han R, Yue Q, Wu R, Li W, Zhao J. New insights into epigenetic modifications in heart failure. Front Biosci. 2017;22:230–47.
Wilk R, Hu J, Blotsky D, Krause HM. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016;30:594–609.
Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–72.
Sun X, Han Q, Luo H, Pan X, Ji Y, Yang Y, Chen H, Wang F, Lai W, Guan X, Zhang Q, Tang Y, Chu J, Yu J, Shou W, Deng Y, Li X. Profiling analysis of long non-coding RNAs in early postnatal mouse hearts. Sci Rep. 2017;7:43485.
Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–40.
Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–86.
Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5.
Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C, Dorn GW. Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci U S A. 2014;111:12264–9.
Zhou X, Zhang W, ** M, Chen J, Xu W, Kong X. LncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8:e2929.
Lv L, Li T, Li X, Xu C, Liu Q, Jiang H, Li Y, Liu Y, Yan H, Huang Q, Zhou Y, Zhang M, Shan H, Liang H. The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Mol Ther Nucleic Acids. 2018;10:387–97.
Liu Y, Zhou D, Li G, Ming X, Tu Y, Tian J, Lu H, Yu B. Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell Physiol Biochem. 2015;35:1986–98.
Rayner KJ, Liu PP. Long noncoding RNAs in the heart: the regulatory roadmap of cardiovascular development and disease. Circ Cardiovasc Genet. 2016;9:101–3.
Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–509.
Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, Boyer LA. A G-rich motif in the lncRNA Braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol Cell. 2016;64:37–50.
Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–20.
Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls hand2 expression and heart development. Nature. 2016;539:433–6.
Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, Zhai M, Huang Y, Yan KW, Dong YH, Ponnusamy M, Shan C, Xu S, Wang Q, Zhang J, Wang K. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29.
Qu X, Du Y, Shu Y, Gao M, Sun F, Luo S, Yang T, Zhan L, Yuan Y, Chu W, Pan Z, Wang Z, Yang B, Lu Y. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep. 2017;7:42657.
Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596.
Zhang G, Sun H, Zhang Y, Zhao H, Fan W, Li J, Lv Y, Song Q, Zhang M, Shi H. Characterization of dysregulated lncRNA-mRNA network based on ceRNA hypothesis to reveal the occurrence and recurrence of myocardial infarction. Cell Death Discov. 2018;4:35.
Zhang Z, Gao W, Long QQ, Zhang J, Li YF, Liu DC, Yan JJ, Yang ZJ, Wang LS. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a chinese population. Sci Rep. 2017;7:7491.
Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, Garg A, Remke J, Zimmer K, Batkai S, Thum T. Inhibition of the cardiac fibroblast-enriched lncRNA MEG3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res. 2017;121:575–83.
Ounzain S, Burdet F, Ibberson M, Pedrazzini T. Discovery and functional characterization of cardiovascular long noncoding RNAs. J Mol Cell Cardiol. 2015;89:17–26.
Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, Dorn GW, Thum T, Heymans S. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol. 2015;12:415–25.
Leti F, DiStefano JK. Long noncoding RNAs as diagnostic and therapeutic targets in type 2 diabetes and related complications. Genes. 2017;8.
Leung A, Natarajan R. Long noncoding RNAs in diabetes and diabetic complications. Antioxid Redox Signal. 2017.
Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12:360–73.
Viereck J, Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res. 2017;120:381–99.
Zhuo C, Jiang R, Lin X, Shao M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget. 2017;8:1429–37.
Li X, Wang H, Yao B, Xu W, Chen J, Zhou X. LncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci Rep. 2016;6:36340.
Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32:473–80.
Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5.
DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–59.
Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci U S A. 1984;81:5523–7.
Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000.
Lustig O, Ariel I, Ilan J, Lev-Lehman E, De-Groot N, Hochberg A. Expression of the imprinted gene H19 in the human fetus. Mol Reprod Dev. 1994;38:239–46.
Milligan L, Antoine E, Bisbal C, Weber M, Brunel C, Forne T, Cathala G. H19 gene expression is up-regulated exclusively by stabilization of the RNA during muscle cell differentiation. Oncogene. 2000;19:5810–6.
Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, VandeBerg JL, McCarrey JR, Consortium S, Renfree MB, Reik W, Dunham I. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–6.
Huang Y, Zheng Y, Jia L, Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/SMAD3/HDAC signaling pathway by deriving miR-675. Stem Cells. 2015;33:3481–92.
Zhang L, Zhou Y, Huang T, Cheng AS, Yu J, Kang W, To KF. The interplay of lncRNA-H19 and its binding partners in physiological process and gastric carcinogenesis. Int J Mol Sci. 2017;18.
Shimizu S, Matsuoka Y, Shinohara Y, Yoneda Y, Tsujimoto Y. Essential role of voltage-dependent anion channel in various forms of apoptosis in mammalian cells. J Cell Biol. 2001;152:237–50.
Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–51.
DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345:78–80.
Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82.
Feng CC, Pandey S, Lin CY, Shen CY, Chang RL, Chang TT, Chen RJ, Viswanadha VP, Lin YM, Huang CY. Cardiac apoptosis induced under high glucose condition involves activation of IGF2r signaling in H9C2 cardiomyoblasts and streptozotocin-induced diabetic rat hearts. Biomed Pharmacother. 2018;97:880–5.
Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, Dandolo L. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–21.
Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3.
Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–65.
Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859:192–9.
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38.
Gu J, **a Z, Luo Y, Jiang X, Qian B, **e H, Zhu JK, **ong L, Zhu J, Wang ZY. Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in arabidopsis thaliana. Nucleic Acids Res. 2018;46:1777–92.
Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014;159:188–99.
Luan W, Li L, Shi Y, Bu X, **a Y, Wang J, Djangmah HS, Liu X, You Y, Xu B. Long non-coding RNA MALAT1 acts as a competing endogenous rna to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7:63901–12.
Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41.
Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB. Chess a. a screen for nuclear transcripts identifies two linked noncoding RNAs associated with sc35 splicing domains. BMC Genomics. 2007;8:39.
Zhang M, Gu H, Xu W, Zhou X. Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int J Cardiol. 2016;203:214–6.
Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–99.
Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34:1249–59.
Zou ZQ, Xu J, Li L, Han YS. Down-regulation of SENCR promotes smooth muscle cells proliferation and migration in db/db mice through up-regulation of Foxo1 and TRPC6. Biomed Pharmacother. 2015;74:35–41.
Riches K, Angelini TG, Mudhar GS, Kaye J, Clark E, Bailey MA, Sohrabi S, Korossis S, Walker PG, Scott DJ, Porter KE. Exploring smooth muscle phenotype and function in a bioreactor model of abdominal aortic aneurysm. J Transl Med. 2013;11:208.
Dorn GW 2nd. LIPCAR: a mitochondrial lnc in the noncoding RNA chain? Circ Res. 2014;114:1548–50.
Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–93.
Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–75.
Aneja A, Tang WH, Bansilal S, Garcia MJ, Farkouh ME. Diabetic cardiomyopathy: insights into pathogenesis, diagnostic challenges, and therapeutic options. Am J Med. 2008;121:748–57.
Seferovic PM, Paulus WJ. Clinical diabetic cardiomyopathy: a two-faced disease with restrictive and dilated phenotypes. Eur Heart J. 2015;36(27):1718 1727a-27c.
Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6.
Holdt LM, Sass K, Gabel G, Bergert H, Thiery J, Teupser D. Expression of chr9p21 genes CDKN2B (p15 ink4b), CDKN2A (p16 ink4a, p14 ARf) and MTAP in human atherosclerotic plaque. Atherosclerosis. 2011;214:264–70.
Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–6.
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–40.
Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-1β-induced cell cycle arrest. Nature. 1994;371:257–61.
Kong Y, Hsieh CH, Alonso LC. ANRIL: a lncRNA at the CDKN2A/bb locus with roles in cancer and metabolic disease. Front Endocrinol. 2018;9:405.
Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74.
McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91.
Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3.
Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53.
Kojima Y, Downing K, Kundu R, Miller C, Dewey F, Lancero H, Raaz U, Perisic L, Hedin U, Schadt E, Maegdefessel L, Quertermous T, Leeper NJ. Cyclin-dependent kinase inhibitor 2b regulates efferocytosis and atherosclerosis. J Clin Invest. 2014;124:1083–97.
Campa D, Pastore M, Gentiluomo M, Talar-Wojnarowska R, Kupcinskas J, Malecka-Panas E, Neoptolemos JP, Niesen W, Vodicka P, Delle Fave G, Bueno-de-Mesquita HB, Gazouli M, Pacetti P, Di Leo M, Ito H, Kluter H, Soucek P, Corbo V, Yamao K, Hosono S, Kaaks R, Vashist Y, Gioffreda D, Strobel O, Shimizu Y, Dijk F, Andriulli A, Ivanauskas A, Bugert P, Tavano F, Vodickova L, Zambon CF, Lovecek M, Landi S, Key TJ, Boggi U, Pezzilli R, Jamroziak K, Mohelnikova-Duchonova B, Mambrini A, Bambi F, Busch O, Pazienza V, Valente R, Theodoropoulos GE, Hackert T, Capurso G, Cavestro GM, Pasquali C, Basso D, Sperti C, Matsuo K, Buchler M, Khaw KT, Izbicki J, Costello E, Katzke V, Michalski C, Stepien A, Rizzato C, Canzian F. Functional single nucleotide polymorphisms within the cyclin-dependent kinase inhibitor 2a/2b region affect pancreatic cancer risk. Oncotarget. 2016;7:57011–20.
Campa D, Capurso G, Pastore M, Talar-Wojnarowska R, Milanetto AC, Landoni L, Maiello E, Lawlor RT, Malecka-Panas E, Funel N, Gazouli M, De Bonis A, Kluter H, Rinzivillo M, Delle Fave G, Hackert T, Landi S, Bugert P, Bambi F, Archibugi L, Scarpa A, Katzke V, Dervenis C, Lico V, Furlanello S, Strobel O, Tavano F, Basso D, Kaaks R, Pasquali C, Gentiluomo M, Rizzato C, Canzian F. Common germline variants within the CDNK2A/2B region affect risk of pancreatic neuroendocrine tumors. Sci Rep. 2016;6:39565.
Rahimi E, Ahmadi A, Boroumand MA, Mohammad Soltani B, Behmanesh M. Association of ANRIL expression with coronary artery disease in type 2 diabetic patients. Cell J. 2018;20:41–5.
Pant T, Dhanasekaran A, Bosnjak ZJ, Ge ZD. Microarray analysis of long noncoding RNAs in the heart and plasma of type 2 diabetic db/db mice. FASEB J. 2018;32:A580.517.
Zur Bruegge J, Einspanier R, Sharbati S. A long journey ahead: long non-coding RNAs in bacterial infections. Front Cell Infect Microbiol. 2017;7:95.
Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–71.
Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–77.
Prabhakar B, Zhong XB, Rasmussen TP. Exploiting long noncoding RNAs as pharmacological targets to modulate epigenetic diseases. Yale J Biol Med. 2017;90:73–86.
Zhou T, Kim Y, MacLeod AR. Targeting long noncoding RNA with antisense oligonucleotide technology as cancer therapeutics. Methods Mol Biol. 2016;1402:199–213.
Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, Taiana E, Agnelli L, Cantafio MEG, Romeo E, Raimondi L, Caracciolo D, Zuccala V, Rossi M, Neri A, Munshi NC, Tagliaferri P, Tassone P. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018.
Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, Maric D, Maison D, Nemir M, Young RA, Schroen B, Gonzalez A, Ounzain S, Pedrazzini T. The long noncoding RNA WISPER controls cardiac fibrosis and remodeling. Sci Transl Med. 2017;9.
Li DY, Busch A, ** H, Chernogubova E, Pelisek J, Karlsson J, Sennblad B, Liu S, Lao S, Hofmann P, Backlund A, Eken SM, Roy J, Eriksson P, Dacken B, Ramanujam D, Dueck A, Engelhardt S, Boon RA, Eckstein HH, Spin JM, Tsao PS, Maegdefessel L. H19 induces abdominal aortic aneurysm development and progression. Circulation. 2018. https://doi.org/10.1161/CIRCULATIONAHA.117.032184.
d'Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, Ling K, Kong L, Ward AJ, Rubin LL, Rigo F, Bennett CF, Sumner CJ. The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron. 2017;93:66–79.
Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, **ao T, Cao Z, Peng J, Yuan P, Brown M, Liu XS, Wei W. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-CAS9 library. Nat Biotechnol. 2016;34:1279–86.
Aparicio-Prat E, Arnan C, Sala I, Bosch N, Guigo R, Johnson R. DECKO: Single-oligo, dual-crispr deletion of genomic elements including long non-coding RNAs. BMC Genomics. 2015;16:846.
Acknowledgements
None.
Funding
This work was supported, in part, by a National Institutes of Health research grant P01GM 066730 (to Dr. Bosnjak) from the United States Public Health Services, Bethesda, Maryland, USA. The funding body had no role in the design of the study, collection, analysis, and interpretation of data, and in writing the manuscript.
Consent of publication
Not applicable.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
ZDG, ZJB, ML, and TP conceived the original idea. TP and JF collected and prepared the literature. ZDG, ZJB, ML, AD, and XB contributed to interpretation of the literature. ZDG and TP reviewed the literature and wrote the original manuscript. All authors read, discussed, and revised the initial manuscript and contributed to the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable-no human projects or tissue.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pant, T., Dhanasekaran, A., Fang, J. et al. Current status and strategies of long noncoding RNA research for diabetic cardiomyopathy. BMC Cardiovasc Disord 18, 197 (2018). https://doi.org/10.1186/s12872-018-0939-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-018-0939-5