Abstract
Background
Bacteria of the order Rickettsiales (Alphaproteobacteria) are obligate intracellular parasites that infect species from virtually every major eukaryotic lineage. Several rickettsial genera harbor species that are significant emerging and re-emerging pathogens of humans. As species of Rickettsiales are associated with an extremely diverse host range, a better understanding of the historical associations between these bacteria and their hosts will provide important information on their evolutionary trajectories and, particularly, their potential emergence as pathogens.
Results
Nine species of Rickettsiales (two in the genus Rickettsia, three in the genus Anaplasma, and four in the genus Ehrlichia) were identified in two species of hard ticks (Dermacentor nuttalli and Hyalomma asiaticum) from two geographic regions in **njiang through genetic analyses of 16S rRNA, gltA, and groEL gene sequences. Notably, two lineages of Ehrlichia and one lineage of Anaplasma were distinct from any known Rickettsiales, suggesting the presence of potentially novel species in ticks in **njiang. Our phylogenetic analyses revealed some topological differences between the phylogenies of the bacteria and their vectors, which led us to marginally reject a model of exclusive bacteria-vector co-divergence.
Conclusions
Ticks are an important natural reservoir of many diverse species of Rickettsiales. In this work, we identified a single tick species that harbors multiple species of Rickettsiales, and uncovered extensive genetic diversity of these bacteria in two tick species from **njiang. Both bacteria-vector co-divergence and cross-species transmission appear to have played important roles in Rickettsiales evolution.
Similar content being viewed by others
Background
Bacteria of the order Rickettsiales are obligate intracellular parasites of eukaryotes. While some symbionts are known (for example, many Wolbachia species), most described species of Rickettsiales are best known as human pathogens that cause several diseases, including rickettsioses, anaplasmosis, and ehrichiosis [1]. Historically, rickettsial agents have been important causes of human morbidity and mortality, including R. prowazekii that caused several million deaths in the USSR [2], and it is estimated that Orientia tsutsugamushi is currently responsible for approximately one million cases of scrub typhus per year [2],[3]. The discovery of new pathogenic species or their associated diseases has attracted attention to the Rickettsiales as pathogens [4]-[8]. As their arthropod vectors often live at high densities and in close proximity to domestic animals and humans, Rickettsiales will continue to pose a risk for transmission to humans. Hence, the identification and characterization of novel Rickettsiales is of importance for both animal and human health.
The number of novel Rickettsiales associated with protists, arthropods, and mammals has increased rapidly through the application of molecular detection and phylogenetics [4]-[6],[9],[10]. Remarkably, analysis of the Trichoplax adhaerens genome also reveals novel species in the order Rickettsiales (for example, [11]). At present, this order contains three established families (Rickettsiaceae, Anaplasmataceae, and Holosporaceae) and one proposed family (Candidatus Midichloriaceae) [8],[11]-[14]. Additionally, some unclassified species warrant further attention to determine their phylogenetic and systematic positions [8],[11]. The intra- and inter-species genetic diversity and evolutionary relationships within genera of Rickettsiales bacteria have been characterized using 16S rRNA gene (rrs) sequences, especially in the case of those bacteria causing animal and human disease [5],[6],[9],[14],[15]. However, relatively little is known about their potential for cross-species transmission and emergence.
Compared with other zoonotic or vector-borne bacteria, Rickettsiales are associated with a more extremely diverse host range, including protists, hydra, annelids, arthropods, vertebrates, and even plants [5],[8],[15],[16]. While some Rickettsiales are specific to particular vectors and hosts [16],[17], others experience host-switching or regularly cycle between different hosts, typically a mammal (e.g. rodents, cattle and humans) and a blood-feeding arthropod (e.g. fleas, mites and ticks) [5],[16],[17]. However, the evolutionary associations between Rickettsiales and their hosts are not well understood [6],[16],[18],[19]. In particular, it is unclear whether Rickettsiales most often evolve by long-term bacteria-host co-divergence or cross-species transmission [20],[21]. As most emerging infectious diseases in humans are caused by spillover from animal hosts or vectors, a better understanding of the evolutionary relationships among Rickettsiales bacteria could provide important information on the likelihood of their emergence as agents of disease.
**njiang (one of five autonomous regions of China) is located in the northwestern part of China, and borders Russia, Mongolia, Kazakhstan, Kyrgyzstan, Tajikistan, Afghanistan, Pakistan and India (Additional file 1: Figure S1) and is one of the nation’s major grazing areas. Several important tick-borne diseases are endemic in **njiang [22]. The main aim of this study was to explore the diversity of Rickettsiales in **njiang, China, where their presence has only previously been shown by serological data [23],[24]. Accordingly, we screened ticks and identified bacteria by sequencing and analyzing three genes; rrs, citrate synthase (gltA), and heat shock protein (groEL). With these data in hand we explored key aspects of Rickettsiales biodiversity and evolution.
Results
Collection of ticks and detection of Rickettsiales bacterial DNA
In the spring of 2011, a total of 2062 adult ticks were collected from domestic animals (sheep and cattle) and grasslands in the border areas of the Bole and Tacheng regions of **njiang Uygur Autonomous Region, China (Additional file 1: Figure S1). The numbers, species, and geographic distributions of the adult ticks collected are shown in Table 1. After morphological examination and sequence analysis of mitochondrial 18S and 12S rDNA sequences as described previously [25], only Dermacentor nuttalli and Hyalomma asiaticum were found in **njiang.
A total of 388 tick pools (1862 ticks) were investigated in this study, 314 of which were from Bole and 74 from Tacheng. PCR was performed to detect Rickettsiales DNA based on rrs. PCR products of the expected size were amplified from 50 tick pools from Bole and 37 from Tacheng. Genetic analyses of these sequences indicated that all products belonged to Rickettisales (see below).
Genetic analysis of bacterial DNA sequences
The rrs, gltA, and groEL gene sequences amplified from the Rickettsiales DNA-positive tick-pool samples were sequenced (sequences are described in detail in Additional file 2: Table S1). Genetic analyses indicated that all sequences recovered from ticks from ** of morphological characters. Syst Biol. 2003, 52: 131-158." href="/article/10.1186/s12862-014-0167-2#ref-CR54" id="ref-link-section-d287326108e2303">54] methods implemented in the Mesquite package [55] to tentatively reconstruct the evolution of habitat among the Rickettsiales by treating “aquatic vs. terrestrial” habitats as discrete character states and map** their occurrence onto the phylogenies.
Analysis of co-divergence events
We tested the hypothesis of bacterial-host co-divergence using the ParaFit method [56] as implemented in the COPYCAT software package [57], which compares the patristic distance matrices derived from the bacteria and vector phylogenies. For this analysis we prepared three tick-only data sets including (i) tick-associated Rickettsia, (ii) Anaplasma, and (iii) Ehrlichia, as well as an overall data set including all Rickettsiales. The bacterial genetic distance matrices were derived from the rrs trees inferred by both BEAST and ML methods, while the vector genetic distance matrices were derived from the 18S rRNA gene trees generated using BEAST as described above. Significance testing was based of 9,999 randomizations of the association matrices. Additionally, to illustrate the association between bacteria (Additional file 6: Table S3, Additional file 8: Table S5) and their vectors, a tanglegram was generated by matching each bacterial species (or group) to their associated vectors using TreeMap 3.0 [58].
Additional files
References
Rickettsial Diseases. 2007, Informa Healthcare, London
Kelly DJ, Richards AL, Temenak JJ, Strickman D, Dasch GA: The past and present threat of rickettsial diseases to military medicine and international public health. Clin Infect Dis. 2002, 34: s145-s169.
Tamura A, Ohashi N, Urakami H, Miyamura S: Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int J Syst Bacteriol. 1995, 45: 589-591.
Zhang L, Liu Y, Ni D, Li Q, Yu Y, Yu XJ, Wan K, Li D, Liang G, Jiang X, **g H, Run J, Luan M, Fu X, Zhang J, Yang W, Wang Y, Dumler JS, Feng Z, Ren J, Xu J: Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA. 2008, 300: 2263-2270.
Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM: Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009, 7: 6-
Merhej V, Raoult D: Rickettsial evolution in the light of comparative genomics. Biol Rev Camb Philos Soc. 2011, 86: 379-405.
Pritt BS, Sloan LM, Johnson DK, Munderloh UG, Paskewitz SM, McElroy KM, McFadden JD, Binnicker MJ, Neitzel DF, Liu G, Nicholson WL, Nelson CM, Franson JJ, Martin SA, Cunningham SA, Steward CR, Bogumill K, Bjorgaard ME, Davis JP, McQuiston JH, Warshauer DM, Wilhelm MP, Patel R, Trivedi VA, Eremeeva ME: Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. N Engl J Med. 2011, 365: 422-429.
Gillespie JJ, Nordberg EK, Azad AF, Sobral BW: Phylogeny and Comparative Genomics: The Shifting Landscape in the Genomics Era. Intracellular Pathogens II: Rickettsiales. Edited by: Azad AF, Palmer GH. 2012, American Society of Microbiology Press, Herndon, Virginia, US
Rar V, Golovljova I: Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011, 11: 1842-1861.
Vaughan JA, Tkach VV, Greiman SE: Neorickettsial endosymbionts of the digenea: diversity, transmission and distribution. Adv Parasitol. 2012, 79: 253-297.
Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW: Bacterial DNA sifted from the Trichoplax adhaerens Animalia: Placozoa genome project reveals a putative rickettsial endosymbiont. Genome Biol Evol. 2013, 5: 621-645.
Brenner DJ, O’Connor SP, Winkler HH, Steigerwalt AG: Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana comb. nov., Bartonella vinsonii comb. nov., Bartonella henselae comb. nov., and Bartonella elizabethae comb. nov., and to remove the family Bartonellaceae from the order Rickettsiales. Int J Syst Bacteriol. 1993, 43: 777-786.
Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR: Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001, 51: 2145-2165.
Montagna M, Sassera D, Epis S, Bazzocchi C, Vannini C, Lo N, Sacchi L, Fukatsu T, Petroni G, Bandi C: “Candidatus Midichloriaceae” fam. nov. Rickettsiales, an ecologically widespread clade of intracellular Alphaproteobacteria. Appl Environ Microbiol. 2013, 79: 3241-3248.
Perlman SJ, Hunter MS, Zchori-Fein E: The emerging diversity of Rickettsia. Proc Biol Sci. 2006, 273 (1598): 2097-2106.
Darby AC, Cho NH, Fuxelius HH, Westberg J, Andersson SG: Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 2007, 23: 511-520.
Renvoisé A, Merhej V, Georgiades K, Raoult D: Intracellular Rickettsiales: Insights into manipulators of eukaryotic cells. Trends Mol Med. 2011, 17: 573-583.
Gillespie JJ, Beier MS, Rahman MS, Ammerman NC, Shallom JM: Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One. 2007, 2: e266-
Walker DH, Ismail N: Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008, 6: 375-386.
Azad AF, Beard CB: Rickettsial pathogens and their arthropod vectors. Emerg Infect Dis. 1984, 4: 179-186.
Toft C, Andersson SG: Evolutionary microbial genomics: Insights into bacterial host adaptation. Nat Rev Genet. 2010, 11: 465-475.
Wu XB, Na RH, Wei SS, Zhu JS, Peng HJ: Distribution of tick-borne diseases in China. Parasit Vectors. 2013, 6: 119-
Chahan B, Jian Z, Xuan X, Sato Y, Kabeya H, Tuchiya K, Itamoto K, Okuda M, Mikami T, Maruyama S, Inokuma H: Serological evidence of infection of Anaplasma and Ehrlichia in domestic animals in **njiang Uygur Autonomous Region area, China. Vet Parasitol. 2005, 134: 273-278.
Sun X, Zhang GL, Liu XM, Zhao Y, Zheng Z: Investigation of tick species and tick-borne pathogens in Hoxud county of **njiang Uyghur Autonomous Region, China. Chin J Vector Biology Control. 2013, 24: 5-7.
Lu X, Lin XD, Wang JB, Qin XC, Tian JH, Guo WP, Fan FN, Shao R, Xu J, Zhang YZ: Molecular survey of hard ticks in endemic areas of tick-borne diseases in China. Ticks Tick Borne Dis. 2013, 4: 288-296.
Garrity GM, Winters M, Kuo AW, Searles DB: Taxonomic Outline of the Prokaryotes. Bergey’s Manual of Systematic Bacteriology, 2nd Edn, Volume II Part C. 2002, Springer-Verlag, New York, US
Mediannikov O, Matsumoto K, Samoylenko I, Drancourt M, Roux V, Rydkina E, Davoust B, Tarasevich I, Brouqui P, Fournier PE: Rickettsia raoultii sp. nov., a spotted fever group rickettsia associated with Dermacentor ticks in Europe and Russia. Int J Syst Evol Microbiol. 2008, 58: 1635-1639.
Beati L, Finidori JP, Raoult D: First isolation of Rickettsia slovaca from Dermacentor marginatus in France. Am J Trop Med Hyg. 1993, 48: 257-268.
Anziani OS, Ewing SA, Barker RW: Experimental transmission of a granulocytic form of the tribe Ehrlichieae by Dermacentor variabilis and Amblyomma americanum to dogs. Am J Vet Res. 1990, 51: 929-931.
Steiert JG, Gilfoy F: Infection rates of Amblyomma americanum and Dermacentor variabilis by Ehrlichia chaffeensis and Ehrlichia ewingii in southwest Missouri. Vector Borne Zoonotic Dis. 2002, 2: 53-60.
Parola P, Inokuma H, Camicas JL, Brouqui P, Raoult D: Detection and identification of spotted fever group Rickettsiae and Ehrlichiae in African ticks. Emerg Infect Dis. 2001, 7: 1014-1017.
Jiang BG, Cao WC, Niu JJ, Wang JX, Li HM, Sun Y, Yang H, Richadus JH, Habbema JD: Detection and identification of Ehrlichia species in Rhipicephalus Boophilus microplus ticks in cattle from **amen, China. Vector Borne Zoonotic Dis. 2011, 11: 325-
Groves MG, Dennis GL, Amyx HL, Huxsoll DL: Transmission of Ehrlichia canis to dogs by ticks Rhipicephalus sanguineus. Am J Vet Res. 1975, 36: 937-940.
Johnson EM, Ewing SA, Barker RW, Fox JC, Crow DW, Kocan KM: Experimental transmission of Ehrlichia canis Rickettsiales: Ehrlichieae by Dermacentor variabilis Acari: Ixodidae. Vet Parasitol. 1998, 74: 277-288.
Jones RT, McCormick KF, Martin AP: Bacterial communities of Bartonella-positive fleas: Diversity and community assembly patterns. Appl Environ Microbiol. 2008, 74: 1667-1670.
Fraune S, Bosch TCG: Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci. 2007, 10: 13146-13151.
Madigan JE, Pusterla N, Johnson E, Chae JS, Pusterla JB, Derock E, Lawler SP: Transmission of Ehrlichia risticii, the agent of Potomac horse fever, using naturally infected aquatic insects and helminth vectors: preliminary report. Equine Vet J. 2000, 32: 275-279.
Huang L, Duan XD, Meng QL, Li R, Zhao QL: Detection of Anaplasma phagocytophilum among sheep in Shihezi, **njiang Uyghur Autonomous Region, China and analysis of its 16S rRNA gene sequences. Chin J Vector Biology Control. 2013, 24: 141-143.
Chen Z, Yang X, Bu F, Yang X, Yang X, Liu J: Ticks Acari: Ixodoidea: Argasidae, Ixodidae of China. Exp Appl Acarol. 2010, 51: 393-404.
Fan MY, Walker DH, Yu SR, Liu QH: Epidemiology and ecology of rickettsial diseases in the People’s Republic of China. Rev Infect Dis. 1987, 9: 823-840.
Fan MY, Jiang YX, Wang LC, Wang JG, Lin YF: Isolation and characterization of tick-borne spotted fever group rickettsia from a patient in **ghe county of **njiang. Chin J Zoonoses. 1985, 2: 9-
Fan MY, Wang JG, Jiang YX, Zong DG, Lenz B: Isolation of a spotted fever group rickettsia from a patient and related ecologic investigations in **njiang Uygur Autonomous Region of China. J Clin Microbiol. 1987, 25: 628-632.
Tian ZC, Liu GY, Shen H, **e JR, Luo J, Tian MY: First report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasit Vectors. 2012, 5: 19-
Raoult D, Lakos A, Fenollar F, Beytout J, Brouqui P, Fournier PE: Spotless rickettsiosis caused by Rickettsia slovaca and associated with Dermacentor ticks. Clin Infect Dis. 2002, 34: 1331-1336.
Ahantarig A, Trinachartvanit W, Baimai V, Grubhoffer L: Hard ticks and their bacterial endosymbionts (or would be pathogens). Folia Microbiol (Praha). 2013, 58: 419-428.
Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier PE, Claverie JM, Raoult D: Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006, 2: e76-
Weisburg WG, Barns SM, Pelletier DA, Lane DJ: 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991, 173: 697-703.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011, 28: 2731-2739.
Moretti S, Wilm A, Higgins DG, Xenarios I, Notredame C: R-Coffee: a web server for accurately aligning noncoding RNA sequences. Nucleic Acids Res. 2008, 36 (Web Server issue): W10-W13.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O: New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010, 59: 307-321.
Huelsenbeck JP, Ronquist F: MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, 17: 754-755.
Drummond AJ, Suchard MA, **e D, Rambaut A: Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012, 29: 1969-1973.
Pagel M: The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst Biol. 1999, 48: 612-622.
Huelsenbeck JP, Nielsen R, Bollback JP: Stochastic map** of morphological characters. Syst Biol. 2003, 52: 131-158.
Maddison WP, Maddison DR: Mesquite: a modular system for evolutionary analysis. Version 2.75. 2011,., [http://mesquiteproject.org]
Legendre P, Desdevises Y, Bazin E: A statistical test for host-parasite coevolution. Syst Biol. 2002, 51: 217-234.
Meier-Kolthoff JP, Auch AF, Huson DH, Göker M: COPYCAT: cophylogenetic analysis tool. Bioinformatics. 2007, 23: 898-900.
Jackson AP, Charleston MA: A cophylogenetic perspective of RNA-virus evolution. Mol Biol Evol. 2004, 21: 45-57.
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grants 81290343, 81273014), State Key Laboratory for Infection Disease Prevention and Control, the Priority Project on Infectious Disease Control and Prevention (Grant 2012ZX10004215), Mega Project of Research on the Prevention and Control of HIV/AIDS, Viral Hepatitis Infectious Diseases (Grant 2011ZX10004-001, 2013ZX10004-101). ECH is supported by a National Health and Medical Research Council Australia Fellowship. JJG acknowledges support from National Institute of Health/National Institute of Allergy and Infectious Diseases grants R01AI017828 and R01AI59118 awarded to Abdu F. Azad (University of Maryland, School of Medicine). JSD is supported in part through grants R01AI44102 and R21AI096062 from the National Institutes of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YZZ conceived the research project; YZZ, YJK, XND, MHC, YX, WMF, YJG, and BP collected the samples, GYZ, YJK, and XPC performed research; YJK, GYZ, and MS analyzed the data; YZZ, YJK, MS, ECH, JJG, and SJD wrote the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12862_2014_167_MOESM1_ESM.jpeg
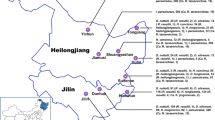
Additional file 1: Figure S1.: Map of sampling locations in **njiang province, China. The Bole and Tacheng regions are labeled by the red dots. (JPEG 254 KB)
12862_2014_167_MOESM3_ESM.pdf
Additional file 3: Figure S2.: Detailed ML phylogenetic trees based on the sequences of Rickettsiales rrs (a, d, g) , gltA (b, e, h), and groEL (c, f, i) genes. The numbers at each branch indicate bootstrap values. (PDF 289 KB)
12862_2014_167_MOESM4_ESM.jpeg
Additional file 4: Figure S3.: Tanglegram of Rickettsiales bacteria and their hosts. The bacterial tree on the left panel of the figure was inferred based on rrs using BEAST and ML (PhyML) methods, while the vector tree on the right panel of the figure was inferred based on 18S rRNA sequences. Each bacterial species (or group) was linked to their associated vectors. In the bacterial tree different genera are distinguished by different colors. The BEAST tree is shown here. (JPEG 791 KB)
12862_2014_167_MOESM7_ESM.doc
Additional file 7: Table S4.: Sequences of the 18S rRNA gene of the vector species used in the phylogenetic analysis. (DOC 54 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kang, YJ., Diao, XN., Zhao, GY. et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol Biol 14, 167 (2014). https://doi.org/10.1186/s12862-014-0167-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-014-0167-2