Abstract
Background
X chromosome inactivation, the mechanism used by mammals to equalise dosage of X-linked genes in XX females relative to XY males, is triggered by chromosome-wide localisation of a cis-acting non-coding RNA, **st. The mechanism of **st RNA spreading and **st-dependent silencing is poorly understood. A large body of evidence indicates that silencing is more efficient on the X chromosome than on autosomes, leading to the idea that the X chromosome has acquired sequences that facilitate propagation of silencing. LINE-1 (L1) repeats are relatively enriched on the X chromosome and have been proposed as candidates for these sequences. To determine the requirements for efficient silencing we have analysed the relationship of chromosome features, including L1 repeats, and the extent of silencing in cell lines carrying inducible **st transgenes located on one of three different autosomes.
Results
Our results show that the organisation of the chromosome into large gene-rich and L1-rich domains is a key determinant of silencing efficiency. Specifically genes located in large gene-rich domains with low L1 density are relatively resistant to **st-mediated silencing whereas genes located in gene-poor domains with high L1 density are silenced more efficiently. These effects are observed shortly after induction of **st RNA expression, suggesting that chromosomal domain organisation influences establishment rather than long-term maintenance of silencing. The X chromosome and some autosomes have only small gene-rich L1-depleted domains and we suggest that this could confer the capacity for relatively efficient chromosome-wide silencing.
Conclusions
This study provides insight into the requirements for efficient **st mediated silencing and specifically identifies organisation of the chromosome into gene-rich L1-depleted and gene-poor L1-dense domains as a major influence on the ability of **st-mediated silencing to be propagated in a continuous manner in cis.
Similar content being viewed by others
Background
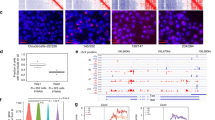
Classical studies on X; autosome rearrangements have demonstrated that X inactivation propagates in cis from a single locus on the X chromosome, the X inactivation centre (** of ** of each transgene (Tg) by DNA FISH, indicating position of nearest BAC probes in Mb. (b) RNA FISH analysis illustrating ** by fluorescence in situ hybridization. Proc Natl Acad Sci USA. 1990, 87: 7757-7761. 10.1073/pnas.87.19.7757." href="/article/10.1186/1756-8935-3-10#ref-CR24" id="ref-link-section-d190910995e749">24]. HL1 domains are often gene poor but are enriched for specific classes of genes, for example olfactory and vomeronasal receptor genes (Additional file 2). LL1 domains on the other hand are highly enriched for genes and also for CpG islands. The extent of HL1 and LL1 domains ranges from <500 Kb through to several Mb. The X chromosome is exceptional in having relatively high L1 and FL-L1 density throughout with only a few gene-rich LL1 domains that are in turn atypically small (Additional file 3).
As illustrated in Figure 3, downregulated genes were located along the entire length of the transgene-bearing chromosomes, chromosome 3, 12 and 17 regardless of their overall length (approximately 160, 120 and 100 Mb, respectively). Their distribution broadly mirrors overall gene density and distance from the transgene integration site does not appear to affect the probability of silencing. These results substantiate RNA FISH data indicating that ** X inactivation. Trends Genet. 2003, 19: 432-438. 10.1016/S0168-9525(03)00177-X." href="/article/10.1186/1756-8935-3-10#ref-CR26" id="ref-link-section-d190910995e913">26]. In the case of Is1ct discontinuous inactivation is at least in part attributable to 'spread and retreat' of X inactivation where chromosome 7 genes are initially silenced and then progressively reactivated [3]. In this study discontinuous inactivation is apparent when ** region (as well as a common Pvu II site in exon 5), using PGK-strain kidney cDNA as PCR template. The RT-PCR products were cloned into pBSX1 by coligation, using the Pvu II and Eco RI sites at exons 5 and 7, respectively, giving pBSX5. Finally, an 8.9 kb Sac II-Pac I exon 1 genomic DNA fragment (also from 129 strain background) was cloned into pBSX5 at the Pac I site, giving pBS** transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997, 94: 3789-3794. 10.1073/pnas.94.8.3789." href="/article/10.1186/1756-8935-3-10#ref-CR35" id="ref-link-section-d190910995e1025">35] as described previously [9]. To test the efficiency of transgene induction doxycycline (1 μg/ml) was added to the medium and 24 h later the cells were screened for transgene expression by RT-PCR across ** data from Ensembl (version 46.36 g), up/downregulated probe sets were defined as those which were mapped to unique locations in the genome and with a FDR ≤ 5%. Downregulated genes were defined as those represented by at least one downregulated probe set and no upregulated probe set at FDR ≤ 5%. The expression value of a downregulated gene was calculated as the average of all associated downregulated probe sets. See Additional file 10 for further details.
Statistical modelling was used to determine whether there was a specific subset of genomic features that explained the observed patterns of gene expression. We employed non-parametric methods in order to model the conditional distribution of gene expression given the genomic features while also determining the importance of each feature and how the salient features interact with each other. Classification and regression trees (CART) models were fit to the experimental data because of their ability to model complex relationships between features, possibly measured on different scales, and deliver compact statistical representations or rules that can be easily visualized and interpreted. Parameter estimation was carried out using the recently developed conditional inference framework for fitting tree-based models based on permutation tests. This framework allowed us to perform feature selection using hypothesis testing procedures. See Additional file 10 for further details.
References
Russell LB: Mammalian X-chromosome action: inactivation limited in spread and region of origin. Science. 1963, 140: 976-978. 10.1126/science.140.3570.976.
Disteche CM, Eicher EM, Latt SA: Late replication in an X-autosome translocation in the mouse: correlation with genetic inactivation and evidence for selective effects during embryogenesis. Proc Natl Acad Sci USA. 1979, 76: 5234-5238. 10.1073/pnas.76.10.5234.
Cattanach BM: Position effect variegation in the mouse. Genet Res. 1974, 23: 291-306. 10.1017/S0016672300014932.
Searle AG, Beechey CV, Evans EP, Kirk M: Two new X-autosome translocations in the mouse. Cytogenet Cell Genet. 1983, 35: 279-292. 10.1159/000131880.
Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, **ng Y, Lawrence J, Willard HF: The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992, 71: 527-542. 10.1016/0092-8674(92)90520-M.
Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S: The product of the mouse **st gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992, 71: 515-526. 10.1016/0092-8674(92)90519-I.
Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N: Requirement for **st in X chromosome inactivation. Nature. 1996, 379: 131-137. 10.1038/379131a0.
Lee JT, Jaenisch R: Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997, 386: 275-279. 10.1038/386275a0.
Wutz A, Jaenisch R: A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000, 5: 695-705. 10.1016/S1097-2765(00)80248-8.
Duthie SM, Nesterova TB, Formstone EJ, Keohane AM, Turner BM, Zakian SM, Brockdorff N: **st RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet. 1999, 8: 195-204. 10.1093/hmg/8.2.195.
Popova BC, Tada T, Takagi N, Brockdorff N, Nesterova TB: Attenuated spread of X-inactivation in an X; autosome translocation. Proc Natl Acad Sci USA. 2006, 103: 7706-7711. 10.1073/pnas.0602021103.
Keohane AM, Barlow AL, Waters J, Bourn D, Turner BM: H4 acetylation, XIST RNA and replication timing are coincident and define x; autosome boundaries in two abnormal X chromosomes. Hum Mol Genet. 1999, 8: 377-383. 10.1093/hmg/8.2.377.
Hall LL, Clemson CM, Byron M, Wydner K, Lawrence JB: Unbalanced X; autosome translocations provide evidence for sequence specificity in the association of XIST RNA with chromatin. Hum Mol Genet. 2002, 11: 3157-3165. 10.1093/hmg/11.25.3157.
White WM, Willard HF, Van Dyke DL, Wolff DJ: The spreading of X inactivation into autosomal material of an x; autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am J Hum Genet. 1998, 63: 20-28. 10.1086/301922.
Gartler SM, Riggs AD: Mammalian X-chromosome inactivation. Annu Rev Genet. 1983, 17: 155-190. 10.1146/annurev.ge.17.120183.001103.
Lyon MF: X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998, 80: 133-137. 10.1159/000014969.
Kazazian HH, Moran JV: The impact of L1 retrotransposons on the human genome. Nat Genet. 1998, 19: 19-24. 10.1038/ng0598-19.
Bailey JA, Carrel L, Chakravarti A, Eichler EE: Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA. 2000, 97: 6634-6639. 10.1073/pnas.97.12.6634.
Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, et al: The DNA sequence of the human X chromosome. Nature. 2005, 434: 325-337. 10.1038/nature03440.
Carrel L, Park C, Tyekucheva S, Dunn J, Chiaromonte F, Makova KD: Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet. 2006, 2: e151-10.1371/journal.pgen.0020151.
Hansen RS: X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Hum Mol Genet. 2003, 12: 2559-2567. 10.1093/hmg/ddg268.
Han JS, Szak ST, Boeke JD: Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004, 429: 268-274. 10.1038/nature02536.
Huntley D, Tang YA, Nesterova TB, Butcher S, Brockdorff N: Genome Environment Browser (GEB): a dynamic browser for visualising high-throughput experimental data in the context of genome features. BMC Bioinformatics. 2008, 9: 501-10.1186/1471-2105-9-501.
Boyle AL, Ballard SG, Ward DC: Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyoty** by fluorescence in situ hybridization. Proc Natl Acad Sci USA. 1990, 87: 7757-7761. 10.1073/pnas.87.19.7757.
Abrusan G, Giordano J, Warburton PE: Analysis of transposon interruptions suggests selection for L1 elements on the X chromosome. PLoS Genet. 2008, 4: e1000172.
Brown CJ, Greally JM: A stain upon the silence: genes esca** X inactivation. Trends Genet. 2003, 19: 432-438. 10.1016/S0168-9525(03)00177-X.
Smith KP, Byron M, Clemson CM, Lawrence JB: Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: enrichment in gene rich bands. Chromosoma. 2004, 113: 324-335. 10.1007/s00412-004-0325-1.
Yu F, Zingler N, Schumann G, Stratling WH: Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001, 29: 4493-4501. 10.1093/nar/29.21.4493.
Yang N, Kazazian HH: L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006, 13: 763-771. 10.1038/nsmb1141.
Chaumeil J, Le Baccon P, Wutz A, Heard E: A novel role for **st RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006, 20: 2223-2237. 10.1101/gad.380906.
Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, Wutz A: SATB1 defines the developmental context for gene silencing by **st in lymphoma and embryonic cells. Dev Cell. 2009, 16: 507-516. 10.1016/j.devcel.2009.03.006.
Federico C, Scavo C, Cantarella CD, Motta S, Saccone S, Bernardi G: Gene-rich and gene-poor chromosomal regions have different locations in the interphase nuclei of cold-blooded vertebrates. Chromosoma. 2006, 115: 123-128. 10.1007/s00412-005-0039-z.
Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, Cremer C, Bult CJ, O'Brien TP: Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006, 174: 27-38. 10.1083/jcb.200603083.
Friedrich G, Soriano P: Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991, 5: 1513-1523. 10.1101/gad.5.9.1513.
Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P: Disruption of overlap** transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997, 94: 3789-3794. 10.1073/pnas.94.8.3789.
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP: Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31: e15-10.1093/nar/gng015.
Smyth GK: Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004, 3: 3.
Benjamini Y, Hochberg Y: Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995, 57: 289-300.
Acknowledgements
We would like to thank the members of the CSC microarray facility for technical assistance and members of the lab for helpful discussion. This work was funded by the MRC UK and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YAT carried out most of the experimental work, contributed to the bioinformatic and statistical analysis and to the study design and manuscript preparation. DH and GM contributed to the bioinformatic and statistical analysis and manuscript preparation. AC contributed to the experimental work and manuscript preparation. TBN and NB contributed to the study design and manuscript preparation. All authors read and approved the final manuscript.
Electronic supplementary material
13072_2010_38_MOESM1_ESM.TIFF
Additional file 1: Examples of L1 domain organisation. (a) Genome Environment Browser (GEB) histogram plot of mouse chromosome 4 illustrating the frequency of genes (green bars), CpG islands (lilac bars), full length L1 (FL-L1) elements (light blue bars), L1 homology sequences (yellow bars) and short interspersed nuclear elements (SINEs, grey bars) within 1-Mb intervals. The 25 Mb regions highlighted by a red box are shown in GEB detailed view in (b). (b) The 25 Mb region 1 (top panel) with boxed areas illustrating small (left) and moderate (right) sized gene-rich domains that are depleted for FL-L1. Note that gene density (green boxes) is higher in these domains. The 25 Mb region 2 (bottom panel) illustrates a large low L1 density (LL1) domain (approximately 22 Mb) flanked on each side by a small high L1 density (HL1) domain. (TIFF 4 MB)
13072_2010_38_MOESM2_ESM.TIFF
Additional file 2: Olfactory receptor (OR) gene clusters localise to high L1 density (HL1) domains. (a) Top panel: Genome Environment Browser (GEB) histogram display for mouse chromosome 9 illustrating one of the largest OR clusters in the mouse genome (marked by a red box), showing high gene density, low CpG island density and high full length L1 (FL-L1) density. Red arrows indicate other gene-rich regions on the chromosome that, as is generally the case, do not show FL-L1 enrichment. The lower panel shows the GEB detailed view of the OR cluster, black dotted lines marking the boundaries. (b) GEB detailed view illustrating a 6-Mb region covering the major histocompatibility complex (MHC) and an OR cluster on mouse chromosome 17 (34.0-40.0 Mb). Boxed areas mark the boundaries of the gene clusters. The OR genes and the MHC are in contrasting HL1 and low L1 density (LL1) domains, respectively. (c) GEB detailed view illustrating a 6-Mb region covering Hist1 histone (H) and vomeronasal receptor (VR) domains on mouse chromosome 13 (19.5-25.5 Mb). Boxed areas mark the boundaries of the gene clusters. Histone gene clusters are L1 depleted whereas VR, like OR, are in an HL1 domain. (d) Scatter plots illustrating L1 density of OR gene clusters in mouse and human (blue). Hist1 histone clusters are included for comparison (red). (TIFF 6 MB)
13072_2010_38_MOESM3_ESM.TIFF
Additional file 3: Unique organisation of high L1 density (HL1) and low L1 density (LL1) domains on the mouse X chromosome. (a) Genome Environment Browser (GEB) histogram display of features on the mouse X chromosome as described for Additional file 1. Boxed 25 Mb regions 1-4 that have relatively high gene density are shown as GEB detailed view in (b). (b) Detailed views of 25 Mb domains. Note that LL1 regions in which genes are concentrated are very small compared with typical autosomes and overall L1 density is high across the entire chromosome. (TIFF 7 MB)
13072_2010_38_MOESM5_ESM.DOC
Additional file 5: Supplementary table 2. File contains Ensembl coordinates (version 46.36 g) for the position of high L1 density (HL1) and low L1 density (LL1) domains on chromosomes 3, 12 and 17. (DOC 438 KB)
13072_2010_38_MOESM6_ESM.TIFF
Additional file 6: Classification trees for downregulated genes on chromosome 17. Classification trees comparing (a) 10% most downregulated genes on chromosome 17 with all other genes. The first split divides the chromosome into two parts, proximal and distal of 35 Mb, based on the distance between the gene and a unique Lx5 element at 9.5 Mb (×82). The left side then shows that for the proximal 35 Mb of the chromosome downregulation is associated with location within a high L1 density (HL1) domain truncated by 500 kb at both ends (×37 >0). For genes distal to 35 Mb downregulation is either associated with location in a low L1 density (LL1) domain less than 1.3 Mb in size (×30), or in larger LL1 domains associated with L1 density downstream of a gene (×25) >3.7%. For trees comparing (b) 30%, (c) 40% and (d) 50% most downregulated genes on chromosome 17 with all other genes, the first split from the top node divides the chromosome into two parts, proximal and distal of approximately 70 Mb, based on the distance between the gene and the nearest full-length (FL) L1_Mus2 element (×44) which can be found at four locations on chromosome 17 (20.7, 31.6, 41.2 and 54.5 Mb). In the proximal 70 Mb of the chromosome, the models suggest that genes less susceptible to silencing are located centrally in large LL1 domains, not associated with large HL1s, and/or surrounded by a low (≤ 12.77%) local L1 density. ×5, overlaps (1) or not (0) with an HL1 domain; ×25, L1 density 100 kb upstream of the gene; ×31, size of HL1 domain with which the gene is associated; ×33 and ×34 whether a gene overlaps (1) or not (0) with a core LL1 domain (LL1 truncated by 250 kb or 500 kb, respectively, at its proximal and distal boundaries). Y-axis denotes probability of a gene being silenced (black fill). (TIFF 492 KB)
13072_2010_38_MOESM7_ESM.TIFF
Additional file 7: Genes located in relatively large low L1 density (LL1) domains on chromosome 3 have a lower probability of being silenced. This effect was seen across the first five deciles, in classification trees comparing (a) 10%, (b) 20%, (c) 30%, (d) 40% and (e) 50% most downregulated genes against all other genes. For every decile, the tree estimated from the data only contained one split, which is either based on a LL1 domain size measure (×30), or on the location of the gene with respect to large LL1 domains (×33 and ×34). Generally, genes located in LL1 domains >1.31 Mb in size or those associated with 'core' LL1 regions (×33 or ×34 >0) have approximately 50% less chance of being silenced. Additional files 4 and 9 give further details on feature definition. (TIFF 565 KB)
13072_2010_38_MOESM8_ESM.TIFF
Additional file 8: Lower local short interspersed nuclear elements (SINE) density may be associated with gene silencing on chromosome 12. Classification trees for chromosome 12 data identified a reciprocal relationship between i**st mediated silencing and SINE density. The effect was only observed when we compared the 10% (a) and 20% (b) most strongly downregulated genes on the chromosome to all other genes. ×26, SINE density in the 100 kb region upstream of the gene. (TIFF 268 KB)
13072_2010_38_MOESM9_ESM.TIFF
Additional file 9: Regression trees for chromosome 12. (a) The first split (×80) measures the distance between the gene to the nearest full length (FL) Lx4B element (which is uniquely located at approximately 6.5 Mb), followed by the second split (×53) which records the orientation of the nearest FL L1 Md_F2 to the gene (1 = upstream of the gene, -1 = downstream). (There are 35 copies of FL L1 Md_F2 on chromosome 12 altogether.) The regression tree suggests that the gene silencing effect would be the strongest for a given gene not more than 18.89 Mb away from the FL Lx4B element, (in other words, in the proximal 25.5 Mb of the chromosome), with the nearest FL L1 Md_F2 element being downstream of the gene. (b) When estimated on a subset of genomic features, the regression tree modelled the gene silencing effect differently. Here the gene silencing effect is the strongest for genes with a relatively low local SINE density (≤ 14.5%) and not associated with the core of a large low L1 density (LL1) domain (×34 = 0). Note that genes located in the centre of a large LL1 domain have a median downregulation level of just 10% (node 4 of the tree), which is a very mild effect. Additional file 4 gives details on genomic feature definition. (TIFF 291 KB)
13072_2010_38_MOESM10_ESM.PDF
Additional file 10: Supplementary methods. File includes supplementary methods for RNA and DNA fluorescence in situ hybridisation (FISH) analysis, immunofluorescence, detailed description of statistical analysis, genomic data analysis and supplementary references. (PDF 110 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Tang, Y.A., Huntley, D., Montana, G. et al. Efficiency of **st-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics & Chromatin 3, 10 (2010). https://doi.org/10.1186/1756-8935-3-10
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-8935-3-10