Abstract
Background
Recent evidence suggests that epithelial cancers, including colorectal cancer are driven by a small sub-population of self-renewing, multi-potent cells termed cancer stem cells (CSCs) which are thought to be responsible for recurrence of cancer. One of the characteristics of CSCs is their ability to form floating spheroids under anchorage-independent conditions in a serum-free defined media. The current investigation was undertaken to examine the role of Wnt/β-catenin pathway in regulating the growth and maintenance of colonospheres. Human colon cancer cells HCT-116 (p53 wild type; K-ras mutant), HCT-116 (p53 null; K-ras mutant) and HT-29 (p53 mutant) were used.
Results
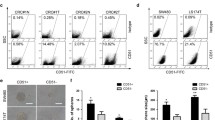
Colonospheres formed in vitro exhibited higher expression of colon CSCs markers LGR5, CD44, CD166 and Musashi-1 along with putative CSC marker EpCAM, compared to the corresponding parental cancer cells and also exhibit the ability to form spheroids under extreme limiting dilution, indicating the predominance of CSCs in colonospheres. Colonospheres formed by HCT-116 cells show over 80% of the cells to be CD44 positive, compared to ≤ 1% in the corresponding parental cells. Additionally, colonospheres showed reduced membrane bound β-catenin but had increased levels of total β-catenin, cyclin-D1 and c-myc and down regulation of axin-1 and phosphorylated β-catenin. Increased expression of β-catenin was associated with a marked transcriptional activation of TCF/LEF. The latter was greatly decreased following down regulation of β-catenin by the corresponding siRNA, leading to a marked reduction in CD44 positive cells as well as colonospheres formation. In contrast, upregulation of c-myc, a down-stream effector of TCF/LEF greatly augmented the formation of colonospheres.
Conclusion
Our data suggest that colonospheres formed by colon cancer cell lines are highly enriched in CSCs and that Wnt/β-catenin pathway plays a critical role in growth and maintenance of colonospheres.
Similar content being viewed by others
Background
A growing body of evidence supports the contention that epithelial cancers including the colorectal cancer are diseases driven by a small set of self renewing cells, termed cancer stem cells (CSC) or cancer-initiating cells, that are distinct from the bulk of the cells in the tumor [1]. Initially identified in hematopoietic tumors, CSCs have now been identified and isolated in a variety of solid tumors that include breast, central nervous system, pancreas, skin, head and neck, colon and prostate [2–7]. CSCs share all the fundamental traits of stem cells-self renewal by asymmetric division, reduced proliferation and differentiation and resistance to apoptosis [8]. CSCs are identified by specific surface epitopes, which in the colon include CD44, CD133 and CD166 [9, 10]. To select putative colorectal CSCs, a promising combination of three markers- EpCAM, CD44 and CD166 was described by Dalerba et al. [11]. Although EpCAM previously being considered as pan-epithelial marker in the normal human colon, its frequent expression in CSCs in breast, colon, pancreas and prostate tumors suggests that this surface epitope could be a putative marker for CSCs, particularly in human colon cancer-derived cell lines [12]. More recently Lgr5, Musashi-1 and aldehyde-dehydrogenase 1 (ALDH-1) have been added to the list of stem cell markers for colon cancer [13–15].
One of the recently reported characteristics of tumor derived CSCs is that they can be grown to form spherical colonies in vitro, when plated in limited numbers under anchorage-independent conditions in a serum-free defined media supplemented with growth factors [9]. With the objectives to promoting in vitro expansions of CSCs, methods have been developed to grow and study them in sphere-forming assays as reported for neurospheres [16, 17], mammospheres [18, 19] and colonospheres [13, 20]. Using this approach, we and others have identified and/expanded colon CSCs by generating colonospheres from colon cancer cell lines [10, Formation of colonospheres The ability of cell lines to form spheres in suspension was evaluated as described by Liu et al. [25], with slight modifications. Briefly, primary colonospheres were generated by incubating the limited number of parental HCT-116 (p53 wild-type; K-ras mutant), HCT-116 (p53 null; K-ras mutant) and HT-29 (p53 mutant; K-ras wild-type) cells at a concentration of 100 cells per 200 μL in serum-free stem cell medium (SCM) containing DMEM/F12 (1:1) supplemented with B27 (Life Technologies, Gaithersburg, MD), 20 ng/ml EGF (Sigma, St Louis, MO), 10 ng/ml fibroblast growth factor (Sigma), and antibiotic-anti-mycotic in 24-well ultra low-attachment plates (Corning Inc, Lowell, MA) for 10 days. The colonospheres formed at the end of the incubation period were centrifuged (1000 rpm), dissociated with 0.05% trypsin/EDTA using a 22 gauge needle and then passed through a 40 μM sieve to obtain single cell suspension, as described by Kakarala et al. [19]. The single-cell suspension derived from colonospheres that have undergone 15 or more serial passages were used for all experiments. For spheroids formation, an equal number of cells from adherent cell lines and colonospheres cells were plated at 200 cells/100 μL SCM in each 96 ultra low-attachment well (Corning Inc, Lowell, MA). The colonospheres formed after 5 days were evaluated for their number and size by light microscopy. Extreme limiting dilution analysis (ELDA) was performed as described by Hu and Smyth [45]. Briefly, single cell suspension obtained from adherent or colonospheres-derived cells were plated at concentration of 1000, 100, 10 cells and 1 cell per 100 μl SCM (24 well for each dilution) in 96-well ultra-low attachment and incubated for 5 days. At the end of 5 days, the number of wells showing formation of colonospheres was counted. The frequency of sphere forming cells in a particular cell type was determined using ELDA webtool at http://bioinf.wehi.edu.au/software/elda. Single cell suspension from parental monolayer cell cultures and colonospheres were subjected to direct immunofluorescence staining followed by flow cytometry analysis according to our standard protocol [22]. Briefly, the cells were harvested and washed with PBS. Half a million cells were suspended in 90 μl of PBS containing 0.5% BSA. After 10 min at room temperature, 10 μl of fluorophore conjugated anti-human CD44 antibody was added and incubated for 30 min in dark at room temperature. The samples were then washed and analyzed using a FACS DiVa (BD, San Jose, CA). The cells stained with mouse IgG2b (isotype-negative control) served as gating control. The proportion of CD44 positive cells was determined on the basis of fluorescence intensity-spectra of CD44-conjugated PE-Cy7 or PerCP-Cy5. Single cell suspension, obtained from parental and colonosphere, was washed in PBS and fixed in 2-4% formaldehyde for 10 min at 37 °C. They were washed and re-suspended in 0.5% BSA-PBS (blocking buffer) for 10 min, subsequently incubated in primary antibodies at appropriate dilution for 1 h at room temperature. After rinsing with incubation buffer, the cells were resuspended in fluorophore-conjugated secondary antibodies diluted in incubation buffer according to the manufacturer's recommendations and incubated for 30 minutes at room temperature. The cells were then resuspended in PBS after washing with the incubation buffer. Appropriate aliquot of cells were mounted on glass slides immediately before examining under fluorescence microscope. Single cell suspension obtained from parental cell lines and colonospheres were washed with PBS (3 times) and stained with Hoechst 33342 or H342 (5 μg/ml, Sigma-Aldrich Inc, St Louis, MO) for 45 minutes at 37°C in HBSS buffer, vortexing gently every 15 min. As a control, a sample was treated with verapamil (Sigma, 50 μM) for ten minutes at room temperature prior to the addition of H342. The stained cells were collected, washed with PBS and resuspended in 3 ml of PBS containing 2 μg/ml of propidium iodide, and subsequently analyzed by flow cytometer-FACS Vantage SE/DiVa SORP (BD Biosciences, San Jose, CA) with all-digital electronics and octagon- and trigon-shaped detector arrays. Excitation of 100 mW at 488 nm was provided by a 177-G argon ion laser (Spectra-Physics, Mountain View, CA) and 200 mW of all-lines UV (351-365 nm) was provided by an Innova 90-5 argon ion laser (Coherent, Palo Alto, CA). Forward and side laser scatter was detected from 488 nm excitation. H342 and propidium iodide fluorescence from UV excitation was split into "blue" and "red" wavelengths by a 505 nm long pass dichroic with a 450/50 bandpass (425-475 nm) filter in front of the "blue" detector and a 630 nm long pass filter in front of the "red" detector. Cell population showing H342 Bright (H342Br) and H342 Low (H342Lo) was determined and the ratio of H342Lo/H3342Br was calculated to evaluate the dye-efflux capacities of the cells. The gating of H342Lo and H342Br cells was based on a verapamil control. Dead cells were gated out based on positive staining with propidium iodide. Changes in cellular growth, an assessment of proliferation, of parental and colonosphere-derived cells were assessed by 3-(4, 5-dimethylthiazol-2yl)-2, 5-diphenyltetrazolium bromide (MTT) assay as described previously [22]. Briefly, 4000 cells were plated per well in 96-well plates in 200 μl medium. At each time point (0, 24, 48, 72, 96, 120 h), 20 μl MTT solution was added to each well and the plate was incubated for 1 h at 37°C. Medium was then aspirated from each well, and 100 μl DMSO was added. The intensity of the color developed, which is the reflection of number of live cells, was measured at a wavelength of 570 nm. All values were compared to the corresponding controls. All assays were performed with 6 replicates. Status of differentiation was determined by measuring the alkaline phosphatase activity using SensoLyte, pNPP alkaline phosphatase assay kit (Anaspec, San Jose, CA) according to the manufacturer's instructions and measuring the absorbance at 405 nm. For the transfection of siRNA into the parental and colonosphere derived cells, Oligofectamine reagent (Invitrogen Corp., Carlsbad, CA) and serum-free Opti-MEM (Invitrogen Corp., Carlsbad, CA) medium was used to prepare transfection complexes according to the manufacturer's instructions. Briefly, single cell suspension of colonospheres was plated in 10 cm tissue culture plates with normal growth medium overnight to achieve 25-30% confluence. Next day the medium was removed, washed twice with serum-free Opti-MEM (Invitrogen Corp., Carlsbad, CA) medium prior to adding the complexes containing non-targeted or β-catenin siRNA (Integrated DNA Technologies Inc., Coralville, IA). After 3 days of transfection, the cells were collected and analyzed for protein expression of β-catenin using Western blot and for colonosphere formation assay using SCM. The activation of transcription factor TCF/LEF was evaluated by using Cignal TCF/LEF reporter assay kit (SA Biosciences, Frederick, MD). The cells were grown to 25-30% confluence as described above and co-transfected with TCF/LEF reporter constructs and either non-targeted or β-catenin siRNAs (Integrated DNA Technologies Inc., Coralville, IA) using SureFECT transfection reagent (SA Biosciences, Frederick, MD) according to manufacturer's instructions. The TCF/LEF reporter used a mixture of an inducible β-catenin-responsive luciferase construct and a constitutively expressing Renilla element (40:1). At the end of 16-24 h incubation period Opti-MEM medium was changed to DMEM 10% FBS for parental cell lines or SCM for colonosphere derived cells. The cells were allowed to grow for another 3 days, collected and analyzed for TCF/LEF activity using a dual-luciferase assay kit (Promega-Biosciences, San Luis Obispo, CA) following the instructions outlined by the manufacturer. Single cell suspension of HCT-116 parental cells was plated in the tissue culture plates to achieve a 90% confluence. Once the 90% confluence is achieved, the adherent cells were transfected using Lipofectamine 2000 and PLUS reagent (Invitrogen Corp., Carlsbad, CA) with plasmid vector-pCMV6-Neo (Origene, Rockville, MD) containing human cDNA clone for c-myc gene or empty plasmid PCMV6-vector in OPTI-MEM medium according to the manufacturer's instruction. After 3 days of transfection, the cells were analyzed for c-myc protein expression by Western blot and re-plated for colonosphere formation assay. Unless otherwise stated, data are expressed as mean ± SD of six observations. Where applicable, the results were analyzed using analysis of variance followed by Fisher protected least significant differences or Scheffé test. p < 0.01 was designated as the level of significance.Extreme limiting dilution analysis
Flow Cytometry Analysis
Immunofluorescence cytochemistry
Hoechst 33342 dye exclusion assay
Determination of cellular growth
Measurement of Alkaline phosphatase activity
β-catenin siRNA transfection
Activation of relative TCF/LEF-Dual Luciferase assay
Overexpression of c-myc gene
Statistical analysis
Authors' information
Shailender S. Kanwar, Ph.D.: Postdoctoral Research Fellow, Department of Internal Medicine and Veterans Affairs Medical Center, Wayne State University, Detroit, MI 48201, USA. E-mail: sskanwar@gmail.com
Yingjie Yu, M.D., Research Assistant Professor, Department of Internal Medicine and Veterans Affairs Medical Center, Wayne State University, Detroit, MI 48201, USA. E-mail: aa5142@wayne.edu
Jyoti Nautiyal, Ph.D.: Postdoctoral Research Fellow, Department of Internal Medicine and Veterans Affairs Medical Center, Wayne State University, Detroit, MI 48201, USA. E-mail: jyotinautiyal@gmail.com
Bhaumik B. Patel, M.D.: Staff Oncologist and Assistant Professor, Department of Internal Medicine, Veterans Affairs Medical Center and Karmanos Cancer Institute, Wayne State University, Detroit, MI 48201, USA. E-mail: bhaumik.patel@va.gov
Adhip P.N. Majumdar, Ph.D., D.Sc.: Professor and Senior Research Career Scientist, Department of Internal Medicine, Veterans Affairs Medical Center and Karmanos Cancer Institute, Wayne State University, Detroit, MI 48201, USA. E-mail: a.majumdar@wayne.edu
References
Jordan CT, Guzman ML, Noble M: Cancer stem cells. N Engl J Med. 2006, 355: 1253-1261. 10.1056/NEJMra061808
Bonnet D, Dick JE: Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997, 3: 730-737. 10.1038/nm0797-730
Jimeno A, Feldmann G, Suárez-Gauthier A, Rasheed Z, Solomon A, Zou G-M, Rubio-Viqueira B, García-García E, López-Ríos F, Matsui W: A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Molecular Cancer Therapeutics. 2009, 8: 310-314. 10.1158/1535-7163.MCT-08-0924
Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ: Efficient tumour formation by single human melanoma cells. Nature. 2008, 456: 593-598. 10.1038/nature07567
Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, Wicha MS, Prince ME: Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head & Neck. 2010, 9999:
Vermeulen L, De Sousa E, Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H: Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010, 12: 468-476. 10.1038/ncb2048
Li T, Su Y, Mei Y, Leng Q, Leng B, Liu Z, Stass SA, Jiang F: ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients/' outcome. Lab Invest. 2009, 90: 234-244. 10.1038/labinvest.2009.127
Dick JE: Stem cell concepts renew cancer research. Blood. 2008, 112: 4793-4807. 10.1182/blood-2008-08-077941
Sukach A, Ivanov E: Formation of spherical colonies as a property of stem cells. Cell and Tissue Biology. 2007, 1: 476-481. 10.1134/S1990519X07060028.
Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF: Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci USA. 2010, 107: 3722-3727. 10.1073/pnas.0915135107
Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM: Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007, 104: 10158-10163. 10.1073/pnas.0703478104
Gires O, Klein CA, Baeuerle PA: On the abundance of EpCAM on cancer stem cells. Nat Rev Cancer. 2009, 9: 143-143. 10.1038/nrc2499-c1
Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP: Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008, 105: 13427-13432. 10.1073/pnas.0805706105
Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, Los M: Cancer stem cell markers in common cancers - therapeutic implications. Trends Mol Med. 2008, 14: 450-460. 10.1016/j.molmed.2008.08.003
Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM: Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009, 69: 3382-3389. 10.1158/0008-5472.CAN-08-4418
Svendsen CN, ter Borg MG, Armstrong RJ, Rosser AE, Chandran S, Ostenfeld T, Caldwell MA: A new method for the rapid and long term growth of human neural precursor cells. J Neurosci Methods. 1998, 85: 141-152. 10.1016/S0165-0270(98)00126-5
Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB: Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63: 5821-5828.
Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, Bundred NJ: Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007, 99: 616-627. 10.1093/jnci/djk133
Kakarala M, Brenner D, Korkaya H, Cheng C, Tazi K, Ginestier C, Liu S, Dontu G, Wicha M: Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Research and Treatment. 2010, 122: 777-785. 10.1007/s10549-009-0612-x
Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R: Identification and expansion of human colon-cancer-initiating cells. Nature. 2007, 445: 111-115. 10.1038/nature05384
Dallas NA, **a L, Fan F, Gray MJ, Gaur P, van Buren G, Samuel S, Kim MP, Lim SJ, Ellis LM: Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009, 69: 1951-1957. 10.1158/0008-5472.CAN-08-2023
Yu Y, Kanwar SS, Patel BB, Nautiyal J, Sarkar FH, Majumdar APN: Elimination of Colon Cancer Stem-Like Cells by the Combination of Curcumin and FOLFOX. Translational Oncology. 2009, 2: 321-328.
Botchkina IL, Rowehl RA, Rivadeneira DE, Karpeh MS, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y, Botchkina GI: Phenotypic Subpopulations of Metastatic Colon Cancer Stem Cells: Genomic Analysis. Cancer Genomics - Proteomics. 2009, 6: 19-29.
Weiswald LB, Guinebretiere JM, Richon S, Bellet D, Saubamea B, Dangles-Marie V: In situ protein expression in tumour spheres: development of an immunostaining protocol for confocal microscopy. BMC Cancer. 2010, 10: 106- 10.1186/1471-2407-10-106
Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS: Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006, 66: 6063-6071. 10.1158/0008-5472.CAN-06-0054
Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS: Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004, 6: R605-615. 10.1186/bcr920
Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E: Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005, 19: 1596-1611. 10.1101/gad.1324905
Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, Clouthier SG, Wicha MS: Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009, 7: e1000121- 10.1371/journal.pbio.1000121
Brabletz S, Schmalhofer O, Brabletz T: Gastrointestinal stem cells in development and cancer. J Pathol. 2009, 217: 307-317. 10.1002/path.2475
Kolligs FT, Bommer G, Goke B: Wnt/beta-catenin/tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion. 2002, 66: 131-144. 10.1159/000066755
Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW: Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997, 275: 1787-1790. 10.1126/science.275.5307.1787
Ricci-Vitiani L, Pagliuca A, Palio E, Zeuner A, De Maria R: Colon cancer stem cells. Gut. 2008, 57: 538-548. 10.1136/gut.2007.127837
Patel VB, Yu Y, Das JK, Patel BB, Majumdar APN: Schlafen-3: A novel regulator of intestinal differentiation. Biochemical and Biophysical Research Communications. 2009, 388: 752-756. 10.1016/j.bbrc.2009.08.094
Fletcher JI, Haber M, Henderson MJ, Norris MD: ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. 2010, 10: 147-156. 10.1038/nrc2789
Scharenberg CW, Harkey MA, Torok-Storb B: The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002, 99: 507-512. 10.1182/blood.V99.2.507
Reya T, Morrison SJ, Clarke MF, Weissman IL: Stem cells, cancer, and cancer stem cells. Nature. 2001, 414: 105-111. 10.1038/35102167
Ishimoto T, Oshima H, Oshima M, Kai K, Torii R, Masuko T, Baba H, Saya H, Nagano O: CD44+ slow-cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Cancer Sci. 2010, 101: 673-678. 10.1111/j.1349-7006.2009.01430.x
Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M: A Tumorigenic Subpopulation with Stem Cell Properties in Melanomas. Cancer Res. 2005, 65: 9328-9337. 10.1158/0008-5472.CAN-05-1343
Lawson DA, Witte ON: Stem cells in prostate cancer initiation and progression. The Journal of Clinical Investigation. 2007, 117: 2044-2050. 10.1172/JCI32810
Levi E, Misra S, Du J, Patel BB, Majumdar AP: Combination of aging and dimethylhydrazine treatment causes an increase in cancer-stem cell population of rat colonic crypts. Biochem Biophys Res Commun. 2009, 385: 430-433. 10.1016/j.bbrc.2009.05.080
Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, Tomlinson IP, Carter NP: Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004, 64: 4817-4825. 10.1158/0008-5472.CAN-04-0328
Willson JK, Bittner GN, Oberley TD, Meisner LF, Weese JL: Cell culture of human colon adenomas and carcinomas. Cancer Res. 1987, 47: 2704-2713.
O'Brien CA, Pollett A, Gallinger S, Dick JE: A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007, 445: 106-110. 10.1038/nature05372
Zenali MJ, Tan D, Li W, Dhingra S, Brown RE: Stemness Characteristics of Fibrolamellar Hepatocellular Carcinoma: Immunohistochemical Analysis with Comparisons to Conventional Hepatocellular Carcinoma. Ann Clin Lab Sci. 2010, 40: 126-134.
Hu Y, Smyth GK: ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009, 347: 70-78. 10.1016/j.jim.2009.06.008
Johnston MD, Edwards CM, Bodmer WF, Maini PK, Chapman SJ: Examples of mathematical modeling: tales from the crypt. Cell Cycle. 2007, 6: 2106-2112. 10.4161/cc.6.17.4649
Christgen M, Ballmaier M, Bruchhardt H, von Wasielewski R, Kreipe H, Lehmann U: Identification of a distinct side population of cancer cells in the Cal-51 human breast carcinoma cell line. Molecular and Cellular Biochemistry. 2007, 306: 201-212. 10.1007/s11010-007-9570-y
Miele L, Takebe N, Ivy SP: The Cancer Stem Cell Hypothesis, Embryonic Signaling Pathways, and Therapeutics: Targeting an Elusive Concept. ASCO EDUCATIONAL BOOK. 2009, 2009: 145-156.
Staal Frank JT, Sen JM: The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. European Journal of Immunology. 2008, 38: 1788-1794. 10.1002/eji.200738118
Pu P, Zhang Z, Kang C, Jiang R, Jia Z, Wang G, Jiang H: Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther. 2009, 16: 351-361. 10.1038/cgt.2008.78
Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M: Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010, 6: 25-36. 10.1016/j.stem.2009.11.013
Ricci-Vitiani L, Fabrizi E, Palio E, De Maria R: Colon cancer stem cells. Journal of Molecular Medicine. 2009, 87: 1097-1104. 10.1007/s00109-009-0518-4
Acknowledgements
This work was supported by grants (APNM) from the National Institutes of Health/National Institute on Aging (AG014343) and the Department of Veterans Affairs.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SSK carried out majority of the experiments and wrote the first draft of the manuscript. He was helped by YY and JN. BBP was involved in the discussion and interpretation of the data. APNM, the principal investigator, was responsible for planning, designing, analysis of the data and overall supervision of the work and final preparation of the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kanwar, S.S., Yu, Y., Nautiyal, J. et al. The Wnt/β-catenin pathway regulates growth and maintenance of colonospheres. Mol Cancer 9, 212 (2010). https://doi.org/10.1186/1476-4598-9-212
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-4598-9-212