Abstract
Background
Thanks to recent high coverage mass-spectrometry studies and reconstructed protein complexes, we are now in an unprecedented position to study the evolution of biological systems. Gene duplications, known to be a major source of innovation in evolution, can now be readily examined in the context of protein complexes.
Results
We observe that paralogs operating in the same complex fulfill different roles: mRNA dosage increase for more than a hundred cytosolic ribosomal proteins, mutually exclusive participation of at least 54 paralogs resulting in alternative forms of complexes, and 24 proteins contributing to bona fide structural growth. Inspection of paralogous proteins participating in two independent complexes shows that an ancient, pre-duplication protein functioned in both multi-protein assemblies and a gene duplication event allowed the respective copies to specialize and split their roles.
Conclusion
Variants with conditionally assembled, paralogous subunits likely have played a role in yeast's adaptation to anaerobic conditions. In a number of cases the gene duplication has given rise to one duplicate that is no longer part of a protein complex and shows an accelerated rate of evolution. Such genes could provide the raw material for the evolution of new functions.
Similar content being viewed by others
Background
Gene duplication can be a major source of innovation in evolution [1], providing redundancy and additional genetic material to build upon and differentiate. In general, eukaryotic genomes contain a large fraction of gene duplicates, with paralogs stemming not only from single gene or segmental duplications, but, in the case of S. cerevisiae, also from a Whole-Genome Duplication event that occurred approximately 100 mln years ago (WGD; [2, 3]). Genomic instability and massive gene loss promptly followed WGD and purged most of the newly formed gene copies from the yeast genome, retaining approximately 10% of them [3]. Today, using multiple genomes of related fungal species with conserved synteny, we can unambiguously identify hundreds of gene pairs as WGD paralogs [4] in addition to normal small scale paralogs.
The identification of paralogs of WGD origin, in conjunction with the wealth of data on physical protein interactions and derived maps of protein complexes, puts us in an unprecedented position to test the fate of nascent duplicated genes and to potentially identify cases of duplication of whole complexes. Recently, it has been shown that, after gene duplication, protein interactions can be conserved [5, 6]. The data suggested that there exists a stepwise pathway of evolution for such functional modules [6], with duplications of homomeric interactions known to have a significant influence on the evolution of genes [5]. Moreover, it is known that gene duplicates can be found less often among the core components of protein complexes compared to sparse regions of protein interaction network [8, 9] and synthetic lethality rate [10], by displaying different phenotypic effects when deleted [11] and occurrence across functional classes (e.g., stress responsive genes, [8]). Musso and colleagues [9] show that nearly half of WGD paralogs co-cluster in the same protein complex. Amoutzias and colleagues [12] indicate that whole genome duplication did not change the dimerization specificities of interacting homologs. Here, we show a much more detailed spectrum of evolutionary and functional fates of higher order protein complex subunits. This integrated overview, enables us to quantify the fates with respect to the duplication type and address questions related to protein specialization (subfunctionalization), as well as the emergence of novel functions related to complexes (neofunctionalization).
Our hypotheses were tested on various types of manually curated data: both complexes from MIPS consortium [13], and those annotated by SGD [14]. To avoid a possible bias introduced by manual curation, we also use computationally derived maps of complexes [15, 16], reconstruction of which was possible owing to recent mass-spectrometry studies [17, 18]. Integration of these datasets allowed us to systematically study the fates of all gene duplicates which are involved in protein complexes.
Results
The fates of duplicate genes in complexes
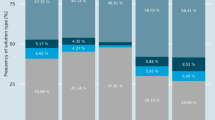
We carried out a systematic analysis of the fate of paralogs in protein complexes. From our first observations it became clear that the cytosolic ribosomal complex dominates the whole spectrum of gene duplications. In order to prevent this single protein complex to dominate our results, we analyze it separately (see Methods). The fates of other paralogs found within complexes fall into two other categories (Figure 1 and 2). Intra-complex paralogs (I) that are formed when both resulting genes remain within the same protein complex, whereas bi-complex paralogs (II) function within two separate complexes. The third class, which we define as overhangs (III), consists of subunits of complexes with a paralog possessing no association to a known protein complex whatsoever. SSD and WGD paralogs are equally divided over intra-complex and overhang classes, but differ with respect to the bi-complex class: many more SSD paralogs are present in two complexes compared to WGD paralogs (Figure 2b). We discuss this observation below.
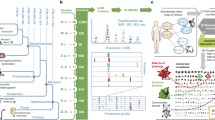
Complex fate of paralogs. a) Gene duplication and subsequent divergence, for cytosolic ribosomal proteins (cRP) followed by homogenizing gene conversion events. b) Impact of duplicated proteins on complexes. Intra-complex duplications include dosage increase, interacting homologs and module variants. Dosage increase requires many components of the complex to duplicate simultaneously (as in the case of cRP and the whole genome duplication). For interacting homologs, the two duplicated proteins become physically subunits of the complex (e.g., homomers turning into heterodimers after the duplication). In module variants only one of the two paralogs is present in the protein complex at a given time. Bi-complex paralogs operate in different protein complexes; two possible evolutionary routes are shown. Overhangs do not aggregate with other proteins in a non-transient manner, while their paralogs do.
The roles of paralogs in protein complexes. a) Shaded areas mark a complex, dashed lines connect paralogs. I) Intra-complex paralogs: when both proteins participate in the same complex; ARG transcription complex includes an intra-complex duplication of genes encoding FUN80 and ARGR1 subunits. II) Bi-complex paralogs: two proteins are involved in different protein complexes; two small complexes are shown: zeta DNA polymerase complex (left) and delta DNA polymerase complex (right). Pair REV3/CDC2 are bi-complex paralogs. III) Overhangs: only one of the paralogs constitutes a subunit of a complex, while its homolog does not aggregate with other proteins in a non-transient manner; Vps4p ATPase transport complex. Here, CHM2 protein (a paralog of DID3) represents an overhang. b) Type of duplication and their contribution to protein complexes: left, whole genome duplication (cytoplasmic ribosomal proteins excluded), and right, small scale duplications. On the pie chart, fractions of all paralog pairs are denoted. Protein complex annotations after SGD consortium.
Intra-complex paralogs: retention is an important fate of paralogs within complexes
We observe a very strong preference for both duplicated proteins to function in the same module. Compared to a null model, where proteins are stochastically reshuffled between complexes, intra-complex paralogs are ~40-fold overrepresented (SGD modules, [14]). This preference is similar, and not statistically different for both duplication types (P = 0.97, chi-square test) and holds for other module definitions, including the computationally derived protein complexes from complex co-purification experiments (see additional file 1, Table S1). Paralog retention within the module is thus an important factor in sha** the map of protein complexes.
Acknowledgements
We would like to thank Ken Wolfe and Gavin Conant for insightful comments. Authors are grateful to Like Fokkens and Jos Boekhorst for discussions, Martin Oti for co-expression dataset, Joanna Parmley for carefully reading the manuscript and Patrick Kemmeren for sharing protein complex data. We would also like to thank anonymous reviewers for their valuable comments. This work was supported by the Netherlands Genomics Initiative (Horizon programme).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
RS and BS designed the study. RS performed the analysis. MH contributed analytical methods. RS and BS wrote the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Szklarczyk, R., Huynen, M.A. & Snel, B. Complex fate of paralogs. BMC Evol Biol 8, 337 (2008). https://doi.org/10.1186/1471-2148-8-337
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-8-337