Abstract
Purpose
To predict potential inhibitors of alpha-enolase to reduce plasminogen binding of Streptococcus pneumoniae (S. pneumoniae) that may lead as an orally active drug. S. pneumoniae remains dominant in causing invasive diseases. Fibrinolytic pathway is a critical factor of S. pneumoniae to invade and progression of disease in the host body. Besides the low mass on the cell surface, alpha-enolase possesses significant plasminogen binding among all exposed proteins.
Methods
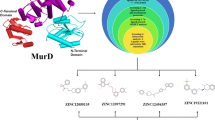
In-silico based drug designing approach was implemented for evaluating potential inhibitors against alpha-enolase based on their binding affinities, energy score and pharmacokinetics. Lipinski’s rule of five (LRo5) and Egan’s (Brain Or IntestinaL EstimateD) BOILED-Egg methods were executed to predict the best ligand for biological systems.
Results
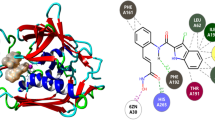
Molecular docking analysis revealed, Sodium (1,5-dihydroxy-2-oxopyrrolidin-3-yl)-hydroxy-dioxidophosphanium (SF-2312) as a promising inhibitor that fabricates finest attractive charges and conventional hydrogen bonds with S. pneumoniae alpha-enolase. Moreover, the pharmacokinetics of SF-2312 predict it as a therapeutic inhibitor for clinical trials. Like SF-2312, phosphono-acetohydroxamate (PhAH) also constructed adequate interactions at the active site of alpha-enolase, but it predicted less favourable than SF-2312 based on binding affinity.
Conclusion
Briefly, SF-2312 and PhAH ligands could inhibit the role of alpha-enolase to restrain plasminogen binding, invasion and progression of S. pneumoniae. As per our investigation and analysis, SF-2312 is the most potent naturally existing inhibitor of S. pneumoniae alpha-enolase in current time.
Graphical abstract







Similar content being viewed by others
Data availability
All data produced during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 5TX:
-
((3 s,5 s)-1,5-Dihydroxy-3-Methyl-2-Oxopyrrolidin-3-Yl)phosphonic acid
- 6BM:
-
[(3 s)-1-Hydroxy-2-Oxopiperidin-3-Yl]phosphonic acid
- BOILED:
-
Brain Or IntestinaL EstimateD
- FSG:
-
(1 s)-1-Fluoro-2-(Hydroxyamino)-2-Oxoethyl]phosphonic acid
- IPD:
-
Invasive pneumococcal disease
- KVM:
-
[(3S)-1-Hydroxy-2,5-Dioxopyrrolidin-3-Yl]phosphonic acid
- LRo5:
-
Lipinski’s rule of five
- NETs:
-
Neutrophil Extracellular Traps
- PhAH:
-
Phosphono-acetohydroxamate
- PLG:
-
Plasminogen
- SF-2312:
-
Sodium (1,5-dihydroxy-2-oxopyrrolidin-3-yl)-hydroxy-dioxidophosphanium
- S. pneumoniae :
-
Streptococcus pneumoniae
- TPSA:
-
Topological polar surface area
References
Lynch JP, Zhanel GG. Streptococcus pneumoniae: epidemiology, risk factors, and strategies for prevention. In Seminars in respiratory and critical care medicine. 2009. © Thieme Medical Publishers.
Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375(9730):1969–87.
Kim L, McGee L, Tomczyk S, Beall B. Biological and epidemiological features of antibiotic-resistant Streptococcus pneumoniae in pre-and post-conjugate vaccine eras: a United States perspective. Clin Microbiol Rev. 2016;29(3):525–52.
Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16(6):355–67.
Bergmann S, Schoenen H, Hammerschmidt S. The interaction between bacterial enolase and plasminogen promotes adherence of Streptococcus pneumoniae to epithelial and endothelial cells. Int J Med Microbiol. 2013;303(8):452–62.
Pancholi V, Fischetti VA. α-Enolase, a novel strong plasmin (ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273(23):14503–15.
Ji H, Wang J, Guo J, Li Y, Lian S, Guo W, et al. Progress in the biological function of alpha-enolase. Anim Nutr. 2016;2(1):12–7.
Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, et al. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8(9):1744–9.
Voss S, Gámez G, Hammerschmidt S. Impact of pneumococcal microbial surface components recognizing adhesive matrix molecules on colonization. Mol Oral Microbiol. 2012;27(4):246–56.
Craig A, Mai J, Cai S, Jeyaseelan S. Neutrophil recruitment to the lungs during bacterial pneumonia. Infect Immun. 2009;77(2):568–75.
Mori Y, Yamaguchi M, Terao Y, Hamada S, Ooshima T, Kawabata S. α-Enolase of Streptococcus pneumoniae induces formation of neutrophil extracellular traps. J Biol Chem. 2012;287(13):10472–81.
Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. 2006;16(4):401–7.
Kolberg J, Aase A, Bergmann S, Herstad TK, Rødal G, Frank R, et al. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology. 2006;152(5):1307–17.
Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server, in The proteomics protocols handbook. 2005, Springer. p. 571–607.
Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6(7):1948–54.
Attique SA, et al. A molecular docking approach to evaluate the pharmacological properties of natural and synthetic treatment candidates for use against hypertension. Int J Environ Res Public Health. 2019;16(6):923.
Buchan DWA, Jones DT. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47(W1):W402–7.
Ehinger S, Schubert WD, Bergmann S, Hammerschmidt S, Heinz DW. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J Mol Biol. 2004;343(4):997–1005.
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–91.
Braga RC, Andrade C. Assessing the performance of 3D pharmacophore models in virtual screening: how good are they? Curr Top Med Chem. 2013;13(9):1127–38.
Warren GL. et al. A critical assessment of docking programs and scoring functions. 2006;49(20):5912–31.
Hu G, et al. Performance evaluation of 2D fingerprint and 3D shape similarity methods in virtual screening. 2012;52(5):1103–13.
Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49(23):6789–801.
Nicholls A. What do we know and when do we know it? J Comput Aided Mol Des. 2008;22(3–4):239–55.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61.
Zalevsky AO, et al. Peptogrid—rescoring function for autodock vina to identify new bioactive molecules from short peptide libraries. Molecules. 2019;24(2):277.
Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–41.
Daina A, Zoete V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11(11):1117–21.
Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1):42717.
MacDougall A, Volynkin V, Saidi R, Poggioli D, Zellner H, Hatton-Ellis E, et al. UniRule: a unified rule resource for automatic annotation in the UniProt knowledgebase. Bioinformatics. 2020.
Edited by Waterbeemd, H.v.d. and B. Testa, Drug bioavailability: estimation of solubility, permeability, absorption and bioavailability. 2009: Wiley-VCH Verlag GmbH.
Edwards MP, Price DA. Chapter 23 - Role of Physicochemical Properties and Ligand Lipophilicity Efficiency in Addressing Drug Safety Risks, in Annual Reports in Medicinal Chemistry, J.E. Macor, Editor. 2010, Academic Press. p. 380–391.
Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. α-Enolase of Streptococcus pneumoniae is a plasmin (ogen)-binding protein displayed on the bacterial cell surface. Mol Microbiol. 2001;40(6):1273–87.
Leonard PG, Satani N, Maxwell D, Lin YH, Hammoudi N, Peng Z, et al. SF2312 is a natural phosphonate inhibitor of enolase. Nat Chem Biol. 2016;12(12):1053–8.
Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–5.
Yang L, Liu Q, Zhang X, Liu X, Zhou B, Chen J, et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature. 2020;583(7814):133–8.
Acknowledgements
The research is supported by FRGS/1 /2017/SKK06iUNISZA/02/1 under Kementerian Pendidikan Malaysia (KPM) and Universiti Sultan Zainal Abidin.
Author information
Authors and Affiliations
Contributions
Conceptualization: Muhammad Hassan and Atif Amin Baig; Methodology: Muhammad Hassan, Atif Amin Baig, Nordin Bin Simbak and Mohammad Amjad Kamal; Software analysis: Muhammad Hassan, Syed Awais Attique, Shafqat Abbas and Muhammad Usman; Validation: Fizza Khan, Sara Zahid, Qurat Ul Ain; Writing – original draft: Muhammad Hassan, Sara Zahid, Qurat Ul Ain; Writing – review & editing: Atif Amin Baig and Hanani Ahmad Yusof.
Corresponding author
Ethics declarations
Conflicts of interest/Competing interests
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hassan, M., Baig, A.A., Attique, S.A. et al. Molecular docking of alpha-enolase to elucidate the promising candidates against Streptococcus pneumoniae infection. DARU J Pharm Sci 29, 73–84 (2021). https://doi.org/10.1007/s40199-020-00384-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40199-020-00384-3