Abstract
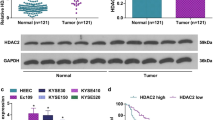
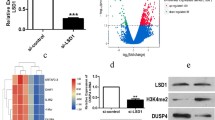
UCHL3 (Ubiquitin carboxyl-terminal hydrolase L3), a member of deubiquitinating enzymes, has been implicated in various cancers. However, the role of UCHL3 in esophageal squamous cell carcinoma (ESCC) remains unknown. In the current study, we aimed to investigate the role of UCHL3 in ESCC growth and migration, and whether UCHL3 could modulate CRY2 methylation through FOXM1. The expression of UCHL3 and CRY2 in ESCC tissues was assessed using qRT-PCR, western blotting and immunohistochemistry (IHC). Cell viability was determined by CCK-8 and colony formation assays. Hoechst 33342 and flow cytometry were used to detect cell apoptosis. Transwell assay was performed to investigate cell migration and invasion. In vivo animal model was used to assess cell tumorigenesis. Methylation-Specific PCR (MSP) was applied to detect CRY2 methylation in the promoter region. The results showed that UCHL3 expression was elevated in ESCC tissues and cells, while CRY2 expression was decreased. UCHL3 silencing inhibited cell viability, invasion, migration and induced cell apoptosis in vitro, repressed tumor growth in vivo, and increased CRY2 expression and decreased FOXM1 expression. In addition, UCHL3 knockdown decreased CRY2 methylation through downregulating FOXM1, leading to an increase in the expression of CRY2. Moreover, CRY2 silencing abolished UCHL3 deficiency-mediated inhibition in cell growth and migration. In summary, this study reveals that knockdown of UCHL3 inhibits ESCC growth and migration by reducing CRY2 methylation through downregulation of FOXM1 expression.







Similar content being viewed by others
References
Ansari MH, Irani S, Edalat H, Amin R, Mohammadi RA. Deregulation of miR-93 and miR-143 in human esophageal cancer. Tumour Biol. 2016;37:3097–103. https://doi.org/10.1007/s13277-015-3987-9.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. https://doi.org/10.3322/caac.21492.
Lin C, Zhang S, Wang Y, Nice E, Guo C, Zhang E, et al. Functional role of a novel long noncoding RNA TTN-AS1 in esophageal squamous cell carcinoma progression and metastasis. Clin Cancer Res. 2018;24:486–98. https://doi.org/10.1158/1078-0432.CCR-17-1851.
Yuequan J, Shifeng C, Bing Z. Prognostic factors and family history for survival of esophageal squamous cell carcinoma patients after surgery. Ann Thorac Surg. 2010;90:908–13. https://doi.org/10.1016/j.athoracsur.2010.05.060.
Cui Y, Chen H, ** R, Cui H, Zhao Y, Xu E, et al. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res. 2020;30:902–13. https://doi.org/10.1038/s41422-020-0333-6.
Hagi T, Kurokawa Y, Takahashi T, Saito T, Yamashita K, Tanaka K, et al. Molecular barcode sequencing for highly sensitive detection of circulating tumor DNA in patients with esophageal squamous cell carcinoma. Oncology. 2020;98:222–9. https://doi.org/10.1159/000504808.
Neutzner M, Neutzner A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012;52:37–50. https://doi.org/10.1042/bse0520037.
Fletcher AJ, Mallery DL, Watkinson RE, Dickson CF, James LC. Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proc Natl Acad Sci USA. 2015;112:10014–9. https://doi.org/10.1073/pnas.1507534112.
Park HB, Kim JW, Baek KH. Regulation of Wnt signaling through ubiquitination and deubiquitination in cancers. Int J Mol Sci. 2020. https://doi.org/10.3390/ijms21113904.
Huang Z, Bao S. Ubiquitination and deubiquitination of REST and its roles in cancers. FEBS Lett. 2012;586:1602–5. https://doi.org/10.1016/j.febslet.2012.04.052.
Wang M, Yu T, Hu L, Cheng Z, Li M. Ubiquitin carboxy-terminal HydrolaseL3 correlates with human sperm count, motility and fertilization. PLoS ONE. 2016;11:e0165198. https://doi.org/10.1371/journal.pone.0165198.
Suzuki M, Setsuie R, Wada K. Ubiquitin carboxyl-terminal hydrolase l3 promotes insulin signaling and adipogenesis. Endocrinology. 2009;150:5230–9. https://doi.org/10.1210/en.2009-0332.
Ouyang L, Yan B, Liu Y, Mao C, Wang M, Liu N, et al. The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor. Signal Transduct Target Ther. 2020;5:78. https://doi.org/10.1038/s41392-020-0181-3.
Li J, Zheng Y, Li X, Dong X, Chen W, Guan Z, et al. UCHL3 promotes proliferation of colorectal cancer cells by regulating SOX12 via AKT/mTOR signaling pathway. Am J Transl Res. 2020;12:6445–54.
Zhang MH, Zhang HH, Du XH, Gao J, Li C, Shi HR, et al. UCHL3 promotes ovarian cancer progression by stabilizing TRAF2 to activate the NF-kappaB pathway. Oncogene. 2020;39:322–33. https://doi.org/10.1038/s41388-019-0987-z.
Song Z, Li J, Zhang L, Deng J, Fang Z, **ang X, et al. UCHL3 promotes pancreatic cancer progression and chemo-resistance through FOXM1 stabilization. Am J Cancer Res. 2019;9:1970–81.
Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. https://doi.org/10.1016/j.cell.2015.03.015.
Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–34. https://doi.org/10.1158/0008-5472.CAN-05-1119.
Mao Y, Fu A, Hoffman AE, Jacobs DI, ** M, Chen K, et al. The circadian gene CRY2 is associated with breast cancer aggressiveness possibly via epigenomic modifications. Tumour Biol. 2015;36:3533–9. https://doi.org/10.1007/s13277-014-2989-3.
Hoffman AE, Zheng T, Stevens RG, Ba Y, Zhang Y, Leaderer D, et al. Clock-cancer connection in non-Hodgkin’s lymphoma: a genetic association study and pathway analysis of the circadian gene cryptochrome 2. Cancer Res. 2009;69:3605–13. https://doi.org/10.1158/0008-5472.CAN-08-4572.
Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009;69:9315–22. https://doi.org/10.1158/0008-5472.CAN-09-0648.
Yu Y, Li Y, Zhou L, Yang G, Wang M, Hong Y. Cryptochrome 2 (CRY2) suppresses proliferation and migration and regulates clock gene network in osteosarcoma cells. Med Sci Monit. 2018;24:3856–62. https://doi.org/10.12659/MSM.908596.
Liu L, Shen H, Wang Y. CRY2 is suppressed by FOXM1 mediated promoter hypermethylation in breast cancer. Biochem Biophys Res Commun. 2017;490:44–50. https://doi.org/10.1016/j.bbrc.2017.06.003.
Zeng K, Wang Z, Ohshima K, Liu Y, Zhang W, Wang L, et al. BRAF V600E mutation correlates with suppressive tumor immune microenvironment and reduced disease-free survival in Langerhans cell histiocytosis. Oncoimmunology. 2016;5:e1185582. https://doi.org/10.1080/2162402X.2016.1185582.
Tian M, Zhu R, Ding F, Liu Z. Ubiquitin-specific peptidase 46 promotes tumor metastasis through stabilizing ENO1 in human esophageal squamous cell carcinoma. Exp Cell Res. 2020;395:112188. https://doi.org/10.1016/j.yexcr.2020.112188.
**g C, Duan Y, Zhou M, Yue K, Zhuo S, Li X, et al. Blockade of deubiquitinating enzyme PSMD14 overcomes chemoresistance in head and neck squamous cell carcinoma by antagonizing E2F1/Akt/SOX2-mediated stemness. Theranostics. 2021;11:2655–69. https://doi.org/10.7150/thno.48375.
Li G, ** X, Zheng J, Jiang N, Shi W. UCH-L3 promotes non-small cell lung cancer proliferation via accelerating cell cycle and inhibiting cell apoptosis. Biotechnol Appl Biochem. 2021;68:165–72. https://doi.org/10.1002/bab.1909.
Fan Y, Hu D, Li D, Ma C, Tang Y, Tao Q, et al. UCHL3 promotes aerobic glycolysis of pancreatic cancer through upregulating LDHA expression. Clin Transl Oncol. 2021. https://doi.org/10.1007/s12094-021-02565-1.
Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. https://doi.org/10.1038/nrc1072.
Rana S, Mahmood S. Circadian rhythm and its role in malignancy. J Circadian Rhythms. 2010;8:3. https://doi.org/10.1186/1740-3391-8-3.
Chu G, Yoshida K, Narahara S, Uchikawa M, Kawamura M, Yamauchi N, et al. Alterations of circadian clockworks during differentiation and apoptosis of rat ovarian cells. Chronobiol Int. 2011;28:477–87. https://doi.org/10.3109/07420528.2011.589933.
Filipski E, King VM, Li X, Granda TG, Mormont MC, Liu X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–7. https://doi.org/10.1093/jnci/94.9.690.
Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33:149–55. https://doi.org/10.1007/s13277-011-0258-2.
Quan M, Wang P, Cui J, Gao Y, **e K. The roles of FOXM1 in pancreatic stem cells and carcinogenesis. Mol Cancer. 2013;12:159. https://doi.org/10.1186/1476-4598-12-159.
Takata A, Takiguchi S, Okada K, Takahashi T, Kurokawa Y, Yamasaki M, et al. Clinicopathological and prognostic significance of FOXM1 expression in esophageal squamous cell carcinoma. Anticancer Res. 2014;34:2427–32.
Zhou Y, Wang Q, Chu L, Dai W, Zhang X, Chen J, et al. FOXM1c promotes oesophageal cancer metastasis by transcriptionally regulating IRF1 expression. Cell Prolif. 2019;52:e12553. https://doi.org/10.1111/cpr.12553.
Song L, Wang X, Feng Z. Overexpression of FOXM1 as a target for malignant progression of esophageal squamous cell carcinoma. Oncol Lett. 2018;15:5910–4. https://doi.org/10.3892/ol.2018.8035.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
JX designed the study, supervised the data collection, JY analyzed the data, interpreted the data, XZ prepared the manuscript for publication and reviewed the draft of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors state that there are no conflicts of interest to disclose.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the standards upheld by the Ethics Committee of Gansu Provincial Cancer Hospital and with those of the 1964 Helsinki Declaration and its later amendments for ethical research involving human subjects (Approval No.2015-11).
Informed consent
Written informed consent was obtained from a legally authorized representative(s) for anonymized patient information to be published in this article.
Research involved in human or animal rights
All animal experiments were approved by the Ethics Committee of Gansu Provincial Cancer Hospital for the use of animals and conducted in accordance with the National Institutes of Health Laboratory Animal Care and Use Guidelines (Apprival No.2018-59).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xue, J., Yi, J. & Zhu, X. Knockdown of UCHL3 inhibits esophageal squamous cell carcinoma progression by reducing CRY2 methylation. Human Cell 35, 528–541 (2022). https://doi.org/10.1007/s13577-021-00660-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00660-7