Abstract
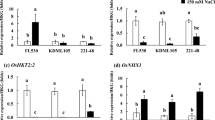
Being a salt-sensitive crop, the growth, development and productivity of rice is severely affected by saline soil. In the present study, Na+ homeostasis, physiological and morphological responses in both the root and leaf tissues of rice (Oryza sativa) seedlings of backcross introgression lines of KDML105 (KD; −/−) × FL530 (+/+) with or without SKC1 and qSt1b QTLs grown under 200 mM NaCl were investigated. Expression levels of OsHKT1;5 in FL530 (+/+) were very low, causing to low Na+ in both the root and leaf tissues, whereas it was up-regulated in KD (−/−), relating to Na+ enrichment. Expression levels of OsHKT2;1 in rice lines, 221-44 (+/+) and 221-48 (+/+) were down-regulated, thus limiting Na+ xylem upload, and store Na+ in the root tissues. OsSOS1 expression level in the leaves of 221-44 (+/+) and 221-58 (+/−) was up-regulated in relation to Na+ efflux. Na+ secretion into vacuoles via OsNHX1 in the leaf tissues was evidently demonstrated in 221-48 (+/+) as a major defense mechanism adopted by the plants to maintain photosynthetic abilities and growth performances, when plants were subjected to salt stress. In addition, the high Fv/Fm ratio in the leaves of 221-58 (+/−) was associated with increased expression level of OsNHX3, in comparison to KD (−/−) seedlings grown under salt stress (diminution by 22.8% over control). Moreover, reduction on growth characters and physiological parameters, viz., shoot fresh weight, shoot height, number of leaves, leaf area, Pn, and accumulation of Na+ in the roots under salt stress were applied as effective indices; thereby classifying FL530 (+/+) and 221-48 (+/+) as salt-tolerant, and 221-44 (+/+), 221-58 (+/−), KD (−/−), 221-3 (+/−), and 221-54 (−/−) as salt-susceptible.





Similar content being viewed by others
Abbreviations
- BILs:
-
Backcross introgression lines
- CRD:
-
Completely randomized design
- CSSLs:
-
Chromosome segment substitution lines
- HKT :
-
High affinity potassium transporter
- MAB:
-
Marker assisted backcrossing
- MS:
-
Murashige and Skoog
- NHX :
-
Na+/H+ exchanger
- NILs:
-
Near isogenic lines
- QTL:
-
Qualitative trait loci
- RIL:
-
Reciprocal inbred line
- SKC1:
-
Shoot K+ concentration
- SOS:
-
Salt overly sensitive
- +/+:
-
Carrying both SKC1 and qSt1b QTLs
- +/−:
-
Carrying only SKC1 QTL
- −/−:
-
Without QTL
References
Ahmadizadeh M, Vispo NA, Calapit-Palao CDO, Pangaan ID, Viña CD, Singh RK (2016) Reproductive stage salinity tolerance in rice: a complex trait to phenotype. Indian J Plant Physiol 21:528–536
Almeida DM, Gregorio GB, Oliveira MM, Saibo NJM (2017) Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol Biol 93:61–77
Arzani A, Ashraf M (2016) Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit Rev Plant Sci 35:146–189
Babu NN, Krishnan SG, Vinod KK, Krishnamurthy SL, Singh VK, Singh MP, Singh R, Ellur RK, Rai V, Bollinedi H, Bhowmick PK, Yadav AK, Nagarajan M, Singh NK, Prabhu KV, Singh AK (2017) Marker aided incorporation of Saltol, a major QTL associated with seedling stage salt tolerance, into Oryza sativa ‘Pusa Basmati 1121’. Front Plant Sci 8:41
Basu S, Giri RK, Benazir I, Kumar S, Rajwanshi R, Dwivedi SK, Kumar G (2017) Comprehensive physiological analyses and reactive oxygen species profiling in drought tolerant rice genotypes under salinity stress. Physiol Mol Biol Plant 23:837–850
Cha-um S, Supaibulwatana K, Kirdmanee C (2006) Water relation, photosynthetic ability and growth of Thai jasmine rice (Oryza sativa L. ssp. indica cv. KDML 105) to salt stress by application of exogenous glycinebetaine and choline. J Agron Crop Sci 192:25–36
Chutimanukul P, Kositsup B, Plaimas K, Buaboocha T, Siangliw M, Too**da T, Comai L, Chadchawan S (2018) Photosynthetic responses and identification of salt tolerance genes in a chromosome segment substitution line of ‘Khao Dawk Mali 105’ rice. Environ Exp Bot 155:497–508
Cotsaftis O, Plett D, Johnson AAT, Walia H, Wilson C, Ismail AM, Close TJ, Tester M, Baumann U (2011) Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol Plant 4:25–41
de Leon TB, Linscombe S, Gregorio G, Subudhi PK (2015) Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front Plant Sci 6:374
Fahad S, Hussain S, Matloob A, Khan FA, Khaliq A, Saud S, Hassan S, Shan D, Khan F, Ullah N, Faiq M, Khan MA, Tareen AK, Khan A, Ullah A, Ullah N, Huang J (2015) Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul 75:391–404
Falakboland Z, Zhou M, Zeng F, Kiani-Pouya A, Shabala L, Shabala S (2017) Plant ionic relation and whole-plant physiological responses to waterlogging, salinity and their combination in barley. Funct Plant Biol 44:941–953
Fu L, Shen Q, Kuang L, YuJ WuD, Zhang G (2018) Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiol Biochem 130:248–257
Fukuda A, Nakamura A, Hara N, Toki S, Tanaka Y (2011) Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233:175–188
Hartley TN, Thomas AS, Maathuis FJ (2020) A role for the OsHKT 2;1 sodium transporter in potassium use efficiency in rice. J Exp Bot 71:699–706
Himabindu Y, Chakradhar T, Reddy MC, Kanygin A, Redding KE, Chandrasekhar T (2016) Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ Exp Bot 124:39–63
Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2; 1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J 26:3003–3014
Horie T, Karahara I, Katsuhara M (2012) Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice 5:11
Hossain GS, Waditee R, Hibino T, Tanaka Y, Takabe T (2006) Root specific expression of Na+/H+ antiporter gene from Synechocystis sp. PCC6803 confers salt tolerance of tobacco plant. Plant Biotechnol 23:275–281
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekee** genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Comm 345:646–651
**g W, Deng P, Cao C, Zhang W (2017) Fine map** of qSKC-1, a major quantitative trait locus for shoot K+ concentration, in rice seedlings grown under salt stress. Breed Sci 67:286–295
Kader MA, Seidei T, Golldack D, Lindberg S (2006) Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J Exp Bot 57:4257–4268
Kanjoo V, Jearakongman S, Punyawaew K, Siangliw JL, Siangliw M, Vanavichit A, Too**da T (2011) Co-location of quantitative trait loci for drought and salinity tolerance in rice. Genom Genet 4:126–138
Kanpawee N, Sanitchon J, Srihaban P, Theerakulpisut P (2012) Genetic diversity analysis of rice cultivars (Oryza sativa L.) differing in salinity tolerance based on RAPD and SSR markers. Electron J Biotechnol 14:4
Kobayashi NI, Yamaji N, Yamamoto H, Okubo K, Ueno H, Costa A, Tanoi K, Matsumura H, Fujii-Kashino M, Horiuchi T, Nayef MA, Shabala S, An G, Ma JF, Horie T (2017) OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J 91:657–670
Lefèvre I, Gratia E, Lutts S (2001) Discrimination between the ionic and osmotic components of salt stress in relation to free polyamine level in rice (Oryza sativa). Plant Sci 161:943–952
Lin HX, Zhu MZZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of shoot and root controlling rice salt tolerance. Theor Appl Genet 108:253–260
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Loggini B, Scartazza A, Brugnoli E, Navari-Izzo F (1999) Antioxidant defense system, pigment composition, and photosynthetic efficiency in two wheat cultivars subjected to drought. Plant Physiol 119:1091–1099
Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51:659–668
Mekawy AMM, Assaha DVM, Yahagi H, Tada Y (2015) Growth, physiological adaptation, and gene expression analysis of two Egyptian rice cultivars under salt stress. Plant Physiol Biochem 87:17–25
Mohammadi-Nejad G, Singh RK, Arzani A, Rezaie AM, Sabouri H, Gregorio GB (2012) Evaluation of salinity tolerance in rice genotypes. Inter J Plant Prod 4:199–207
Moradi F, Ismail AM (2007) Response of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot 99:1161–1173
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, James RA, Gilliham M, Flowers TJ, Colmer TD (2016) Tissue tolerance: an essential but elusive trait for salt-tolerant crops. Funct Plant Biol 43:1103–1113
Nounjan N, Siangliw JL, Too**da T, Chadchawan S, Theerakulpisut P (2016) Salt-responsive mechanisms in chromosome segment substitution lines of rice (Oryza sativa L. cv. KDML105). Plant Physiol Biochem 103:96–105
Omisun T, Sahoo S, Saha B, Panda SK (2018) Relative tolerance of rice cultivars native to North East India: a physiological, biochemical and molecular perspective. Protoplasma 255:193–202
Parihar P, Singh S, Singh R, Singh V, Prasad SM (2015) Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res 22:4056–4075
Punyawaew K, Suriya-Arunroj D, Siangliw M, Thida M, Lanceras-Siangliw J, Fukai S, Too**da T (2016) Thai jasmine rice cultivar KDML105 carrying Saltol QTL exhibiting salinity tolerance at seedling stage. Mol Breed 36:150
Rahman MA, Thomson MJ, Shah-E-Alam M, de Ocampo M, Egdane J, Ismail A (2016) Exploring novel genetic sources of salinity tolerance in rice through molecular and physiological characterization. Ann Bot 117:1083–1097
Ren Z-H, Gao J-P, Li L-G, Cai X-L, Huang W, Chao D-Y, Zhu M-Z, Wang Z-Y, Luan S, Lin H-X (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146
Senadheera P, Singh RK, Maathuis FJM (2009) Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. J Exp Bot 60:2553–2563
Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14:465–477
Siangliw M, Phatanathara A, Too**da T, Theerakulpisut P, Fukai S, Vanavichit A (2014) QTL and candidate gene identification for salt tolerance and Na/K ratio at seedling stage under modified soil and hydroponic conditions in rice (Oryza sativa L.). In: The 4th international rice congress (IRC2014), 27 October–1 November 2014, Bangkok International Trade and Exhibition Centre (BITEC), Bangkok, Thailand
Singh V, Singh AP, Bhadoria J, Giri J, Singh J, Vneeth TV, Sharma PC (2018) Differential expression of salt-responsive genes to salinity stress in salt-tolerant and salt-sensitive rice (Oryza sativa L.) at seedling stage. Protoplasma 255:1667–1681
Suriya-Arunroj D, Supapoj N, Vanavichit A, Too**da T (2005) Screening and selection for physiological characters contributing to salinity tolerance in rice. Kasetsart J 39:174–185
Tanaka K, Ohta K, Haddad PR, Fritz JS, Lee KP, Hasebe K, Ieuji A, Miyanaga A (1999) Acid-rain monitoring in East Asia with a portable-type ion-exclusion-cation-exchange chromatographic analyzer. J Chromatog 850:311–317
Theerawitaya C, Yamada N, Samphumphuang T, Cha-um S, Takabe T, Kirdmanee C (2015) Evaluation of Na+ enrichment and expression of some carbohydrate related genes in indica rice seedlings under salt stress. Plant Omics 8:130–140
Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumward E, Seraj ZI, Singh RK, Gregorio GB, Ismail AM (2010) Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3:148–160
Vanavichit A, Kamolsukyeunyoung W, Siangliw M, Siangliw JL, Traprab S, Ruengphayak S, Chaichoompu E, Saensuk C, Phuvanartnarubal E, Too**da T, Trakoonrung S (2018) Thai Hom Mali rice: origin and breeding for subsistence rainfed lowland rice system. Rice 11:20
Walia H, Wilson A, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Cui X, Close TJ (2015) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139:822–835
Wang H, Zhang M, Guo R, Shi D, Liu B, Lin X, Yang C (2012) Effect of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol 12:194
Wu H (2018) Plant salt tolerance and Na+ sensing and transport. Crop J 6:215–225
Zhang Y, Fang J, Wu X, Dong L (2018) Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol 18:375
Zheng H, Zhao H, Liu H, Wang J, Zou D (2015) QTL analysis of Na+ and K+ concentrations in shoots and roots under NaCl stress based on linkage and association analysis in japonica rice. Euphytica 201:109–121
Zhu M, Zhou M, Shabala L, Shabala S (2017) Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant Cell Environ 40:1009–1020
Acknowledgements
The authors would like to sincerely thank National Science and Technology Development Agency (NSTDA) for funding support (Grant number P-17-50079) and post-doctoral fellowship for CT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Theerawitaya, C., Samphumphuang, T., Tisarum, R. et al. Transcriptional expression of Na+ homeostasis-related genes and physiological responses of rice seedlings under salt stress. J. Plant Biochem. Biotechnol. 30, 81–91 (2021). https://doi.org/10.1007/s13562-020-00573-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-020-00573-w