Abstract
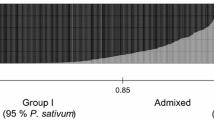
Genetic diversity, population structure and genome-wide marker-trait association analysis was conducted for the USDA pea (Pisum sativum L.) core collection. The core collection contained 285 accessions with diverse phenotypes and geographic origins. The 137 DNA markers included 102 polymorphic fragments amplified by 15 microsatellite primer pairs, 36 RAPD loci and one SCAR (sequence characterized amplified region) marker. The 49 phenotypic traits fall into the categories of seed macro- and micro-nutrients, disease resistance, agronomic traits and seed characteristics. Genetic diversity, population structure and marker-trait association were analyzed with the software packages PowerMarker, STUCTURE and TASSEL, respectively. A great amount of variation was revealed by the DNA markers at the molecular level. Identified were three sub-populations that constituted 56.1%, 13.0% and 30.9%, respectively, of the USDA Pisum core collection. The first sub-population is comprised of all cultivated pea varieties and landraces; the second of wild P. sativum ssp. elatius and abyssinicum and the accessions from the Asian highland (Afghanistan, India, Pakistan, China and Nepal); while the third is an admixture containing alleles from the first and second sub-populations. This structure was achieved using a stringent cutoff point of 15% admixture (q-value 85%) of the collection. Significant marker-trait associations were identified among certain markers with eight mineral nutrient concentrations in seed and other important phenotypic traits. Fifteen pairs of associations were at the significant levels of P ≤ 0.01 when tested using the three statistical models. These markers will be useful in marker-assisted selection to breed pea cultivars with desirable agronomic traits and end-user qualities.
Similar content being viewed by others
References
Abdurakhmonov IY and Abdukarimov A (2008) Application of association map** to understanding the genetic diversity of plant germplasm resources. Int. J. Plant Genomics 574927.
Baranger A, Aubert G, Arnau G, Lain AL, Deniot G, Potier J, Weinachter C, Lejeune-Hénaut I, Lallemand J, and Burstin J (2004) Genetic diversity within Pisum sativum using protein- and PCR-based markers. Theor. Appl. Genet. 108: 1309–1321.
Botstein D, White RL, Skolnick M, and Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Amer. J. Human Genet. 32: 314–331.
Bordat A, Savois V, Nicolas M, Salse J, Chauveau A, Bourgeois M, Potier J, Houtin H, Rond C, Murat F, Marget P, Aubert G and Burstin J (2011) Translational genomics in legumes allowed placing in silico 5460 unigenes on the pea functional map and identified candidate genes in Pisum sativum L. Genes Genom. Genet. 1: 93–103.
Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti AK, Woodward JE, May GD, et al. (2011) Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. USA 108: E864–E870.
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, and Buckler ES (2007) TASSEL: software for association map** of complex traits in diverse samples. Bioinformatics 23: 2633–2635.
Bretting PK and Widrlechner MP (1995) Genetic markers and plant genetic resource management. In Plant Breeding Reviews, J. Janick, ed., John Wiley & Sons, New York, NY, pp. 11–86.
Brown AH (1989) Core collections: a practical approach to genetic resources management. Genome 31: 818–824.
Brown AH and Spillane C (1999) Implementing core collections - principles, procedures, progress, problems and promise. In Core collections for today and tomorrow, R.C. Johnson and T. Hodgkin, eds., Crop Science Society of America, Madison, WI, pp. 1–10.
Burstin J, Deniot G, Potier J, Weinachter C, Aubert G, and Barranger A (2001) Microsatellite polymorphism in Pisum sativum. Plant Breed. 120: 311–317.
Charcosset A and Moreau L (2004) Use of molecular markers for the development of new cultivars and the evaluation of genetic diversity. Euphytica 137: 81–94.
Chavarriaga-Aguirre P, Maya MM, Tohme J, Duque MC, Iglesias C, Bonierbale MW, Kresovich S, and Kochert G (1999) Using microsatellites, isozymes and AFLPs to evaluate genetic diversity and redundancy in the cassava core collection and to assess the usefulness of DNA-based markers to maintain germplasm collections. Mol. Breed. 5: 263–273.
Choudhury PR, Tanveer H, and Dixit GP (2007) Identification and detection of genetic relatedness among important varieties of pea (Pisum sativum L.) grown in India. Genetica 130: 183–191.
Coyne CJ, Brown AF, Timmerman-Vaughn GM, McPhee K, and Grusak MA (2005a) USDA-ARS refined pea core collection for 26 qualitative traits. Pisum Genet. 37: 3–6.
Coyne CJ, Grusak MA, Razai L, and Baik BK (2005b) Variation for pea seed protein concentration in the USDA Pisum core collection. Pisum Genet. 37: 5–9.
Deulvot C, Charrel H, Amandine M, Jacquin F, Donnadieu C, Lejeune-Hénaut I, Burstin J, and Aubert G (2010) High-multiplexed SNP genoty** for genetic map** and germplasm diversity studies in pea. BMC Genomics 11: 468.
Dribnokhodova OP and Gostimsky SA (2009) Allele Polymorphism of Microsatellite Loci in Pea Pisum sativum L. Lines, Varieties, and Mutants. Rus. J. Genet. 45: 788–793.
Ellis THN, Poyser SJ, Knox MR, Vershinin AV, and Ambrose MJ (1998) Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol. Gen. Genet. 260: 9–19.
Esposito MA, Martin EA, Cravero VP, and Cointry E (2007) Characterization of pea accessions by SRAP’s markers. Scientia Hort. 113: 329–335.
Evanno G, Regnaut S, and Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620.
Falush D, Stephens M, and Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Falush D, Stephens M, and Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7: 574–578.
Flint-Garcia SA, Thornsberry JM, and Buckler ES (2003) Structure of linkage disequilibrium in plants. Ann. Rev. Plant Biol. 54: 357–374.
Food and Agricultural Organization of the United Nations (2011) http://www.fao.org/
Ford R, Le Roux K, Itman C, Brouwer JB, and Taylor PWJ (2002) Diversity analysis and genoty** in Pisum with sequence tagged microsatellite site (STMS) primers. Euphytica 124: 397–405.
Franssen SU, Shrestha RP, Bräutigam A, Bornberg-Bauer E, and Weber APM (2011) Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing BMC Genomics 12: 227.
Frary A, Nesbitt TC, Frary A, Grandillo S, van der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, and Tanksley SD (2000) fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88.
Gebhardt C, Ballvora A, Walkemeier B, Oberhagemann P, and Schüler K (2004) Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol. Breed. 13: 93–102.
Ghafoor A, Ahmad Z, and Anwar R (2005) Genetic diversity in Pisum sativum and a strategy for indigenous biodiversity conservation. Pak. J. Bot. 37: 71–77.
Gilpin BJ, McCallum JA, Frew TJ, and Timmerman-Vaughan GM (1997) A linkage map of the pea (Pisum sativum L.) genome containing cloned sequences of known function and expressed sequence tags (ESTs). Theor. Appl. Genet. 95: 1289–1299.
Glaszmann JC, Kilian B, HD Upadhyaya HD, and Varshney RK (2010) Accessing genetic diversity for crop improvement. Curr. Opin. Plant Biol. 13: 167–173.
Grünwald NJ, Coffman VA, and Kraft JM (2003) Sources of partial resistance to Fusarium root rot in the Pisum core collection. Plant Disease 87: 1197–1200.
Grusak MA, Burgett CL, Knewtson SJB, Lopéz-Millán A-F, Ellis DR, Li C-M, Musetti VM, and Blair MW (2004) Novel approaches to improve legume seed mineral nutrition. Proceedings of the 5th AEP-2nd ICLGG Conference: pp. 37–38.
Hagenblad J and Nordborg M (2002) Sequence variation and haplotype structure surrounding the flowering time locus FRI in Arabidopsis thaliana. Genetics 161: 289–298.
Hardy OJ and Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2:618–620.
Hoey BK, Crowe KR, Jones VM, and Polans NO (1996) A phylogenetic analysis of Pisum based on morphological characters, and allozyme and RAPD markers. Theor. Appl. Genet. 92: 92–100.
Hubisz MJ, Falush D, Stephens M, and Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 9: 1322–1332.
Jermyn WA and Slinkard AE (1977) Variability of percent protein and its relationship to seed yield and seed shape in peas. Legume Res. 1: 33–37.
**g R, Johnson R, Seres A, Kiss G, Ambrose MJ, Knox MR, Ellis TH, and Flavell AJ (2007) Gene-based sequence diversity analysis of field pea (Pisum). Genetics 177: 2263–2275.
**g R, Knox MR, Lee JM, Vershinin AV, Ambrose M, Ellis TH, and Flavell AJ (2005) Insertional polymorphism and antiquity of PDR1 retrotransposon insertions in Pisum species. Genetics 171: 741–752.
**g R, Vershinin A, Grzebyta J, Shaw P, Smykal P, Marshall D, Ambrose MJ, Ellis TH, and Flavell AJ (2010) The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol. Biol. 10: 44.
Kosterin OE, Zaytseva OO, Bogdanova VS, and Ambrose MJ (2010) New data on three molecular markers from different cellular genomes in Mediterranean accessions reveal new insights into phylogeography of Pisum sativum L. subsp. elatius (Bieb.) Schmalh. Genet. Res. Crop Evol. 57: 733–739.
Kraft JM, Dunne B, Goulden D, and Armstrong S (1998) A search for resistance in peas to Mycosphaerella pinodes. Plant Disease 82: 251–253.
Laucou V, Haurogn K, Ellis N, Rameau C (1998) Genetic map** in pea. 1. RAPD-based genetic linkage map of Pisum sativum. Theor. Appl. Genet. 97: 905–915.
Lazaro A and Aguinagalde I (2006) Genetic variation among Spanish pea landraces revealed by inter simple sequence repeat (ISSR) markers: its application to establish a core collection. J. Agri. Sci. 144: 53–61.
Le Clerc V, Cadot V, Canadas M, Lallemand J, Guerin D, and Boulineau F (2006) Indicators to assess temporal genetic diversity in the French Catalogue: no losses for maize and peas. Theor. Appl. Genet. 113: 1197–1209.
Lee D, Ellis THN, Turner L, Hellens RP, and Cleary WG (1990) A copia-like element in Pisum demonstrates the uses of dispersed repeated sequences in genetic analysis. Plant Mol. Biol. 15: 707–722.
Liu K and Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129.
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, and Lejeune-Hénaut I (2005) Microsatellite marker polymorphism and map** in pea (Pisum sativum L.). Theor. Appl. Genet. 111: 1022–1031.
Lu J, Knox MR, Ambrose MJ, Brown JKM, and Ellis THN (1996) Comparative analysis of genetic diversity in pea assessed by RFLP-and PCR-based methods. Theor. Appl. Genet. 93: 1103–1111.
Malvick DK and Percich JA (1999) Identification of Pisum sativum germ plasm with resistance to root rot caused by multiple strains of Aphanomyces euteiches. Plant Disease 83: 51–54.
Malysheva-Otto LV, Ganal MW, and Roder MS (2006) Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC Genetics 7: 6.
Martín-Sanz A, Caminero C, **g R, Flavell AJ, and Pérez de la Vega M (2011) Genetic diversity among Spanish pea (Pisum sativum L.) landraces, pea cultivars and the world Pisum sp. core collection assessed by retrotransposon-based insertion polymophisms (RBIPs). Span. J. Agri. Res. 9: 166–78.
Mazzucato A, Papa R, Bitocchi E, Mosconi P, Nanni L, Negri V, Picarella ME, Siligato F, Soressi GP, Tiranti B, and Veronesi F (2008) Genetic diversity, structure and marker-trait associations in a collection of Italian tomato (Solanum lycopersicum L.) landraces. Theor. Appl. Genet. 116: 657–669.
McPhee K (2005) Variation for seedling root architecture in the core collection of pea germplasm. Crop Sci. 45: 1758–1763.
McPhee KE and Muehlbauer FJ (1999) Stem strength in the core collection of Pisum germplasm. Pisum Genet. 31: 21–23.
McPhee KE and Muehlbauer FJ (2001) Biomass production and related characters in the core collection of Pisum germplasm. Genet. Res. Crop Evol. 48: 195–203.
McPhee KE, Tullu A, Kraft JM, and Muehlbauer FJ (1999) Resistance to Fusarium wilt race 2 in the Pisum core collection. J. Amer. Soc. Hort. Sci. 124: 28–31.
Murray MG and Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl. Acids Res. 8: 4321–4325.
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, and Buckler ES (2009) Association map**: critical considerations shift from genoty** to experimental design. Plant Cell 21: 2194–2202.
Neumann K, Kobiljski B, Denčić S, Varshney RK, and Börner A (2011) Genome-wide association map**: a case study in bread wheat (Triticum aestivum L.). Mol. Breed. 27: 37–58.
Nisar M, Ghafoor A, Khan MR, and Subhan M (2009) Genetic similarity of Pakistan pea (Pisum sativum L.) germplasm with world collection using cluster analysis and Jaccard’s similarity index. J. Chem. Soc. Pak. 31: 138–144.
Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, and Oefner PJ (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193.
Okubara PA, Keller KE, McClendon MT, McPhee KE, Inglis DA, and Coyne CJ (2005) Y15_999Fw, a dominant SCAR marker linked to the Fusarium wilt race 1 (Fw) resistance gene in pea. Pisum Genet. 37: 32–35.
Pearce SR, Knox M, Ellis THN, Flavell AJ, and Kumar A (2000) Pea Ty1-copia group retrotransposons: transpositional activity and use as markers to study genetic diversity in Pisum. Mol. Gen. Genet. 263: 898–907.
Pilet-Nayel M, Muehlbauer F, McGee R, Kraft J, Baranger A, and Coyne C (2002) Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor. Appl. Genet. 106: 28–39.
Pritchard JK, Stephens M, and Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945.
Rafalski JA (2010) Association genetics in crop improvement. Curr. Opin. Plant Biol. 13: 174–180.
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, and Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484.
Samec P, Pošvec Z, Stejskal J, Našinec V, and Griga M (1998) Cultivar identification and relationships in Pisum sativum L. based on RAPD and isoenzymes. Biologia Plantarum 41: 39–48.
Schoen DJ and Brown AH (1993) Conservation of allelic richness in wild crop relatives is aided by assessment of genetic markers. Proc. Natl. Acad. Sci. USA 90: 10623–10627.
Simioniuc D, Uptmoor R, Friedt W, Ordon F, and Swiecicki W (2002) Genetic diversity and relationships among pea cultivars revealed by RAPDs and AFLPs. Plant Breed. 121: 429–435.
Simon CJ and Hannan RM (1995) Development and use of core subsets of cool-season food legume germplasm collections. HortSci. 30: 907.
Smith JSC and Smith OS (1992) Fingerprinting crop varieties. Ad. Agronomy 47: 3.
Smýkal P, Kenicer G, Flavell AJ, Corander J, Kosterin O, Redden RJ, Ford R, Coyne CJ, Maxted N, Ambrose MJ, and Ellis THN (2011) Phylogeny, phylogeography and genetic diversity of Pisum genus. Plant Genet. Res. 9: 4–18.
Smýkal P, Hýbl M, Corander J, Jarkovsk J, Flavell AJ, and Griga M (2008) Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theor. Appl. Genet. 117: 413–424.
Stich B, Melchinger AE, Frisch M, Maurer HP, Heckenberger M, and Reif JC (2005) Linkage disequilibrium in European elite maize germplasm investigated with SSRs. Theor. Appl. Genet. 111: 723–730.
Tamura K, Dudley J, Nei M, and Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599.
Tanksley SD and McCouch SR (1997) Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild. Science 277: 1063–1066.
Tar’an B, Zhang C, Warkentin T, Tullu A, and Vandenberg A (2005) Genetic diversity among varieties and wild species accessions of pea (Pisum sativum L.) based on molecular markers, and morphological and physiological characters. Genome 48: 257–272.
Tedford EC and Inglis DA (1999) Evaluation of legumes common to the Pacific northwest as hosts for the pea cyst nematode, Heterodera goettingiana. J. Nematol. 31: 155–163.
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, and Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166.
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, and Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289.
Vershinin AV, Allnutt TR, Knox MR, Ambrose MJ, and Ellis TH (2003) Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Mol. Biol. Evol. 20: 2067.
Weiguo Z, Eun-** P, Jong-Wook C, Yong-** P, Ill-Min C, Joung-Kuk A, and Gwang-Ho K (2009) Association analysis of the amino acid contents in rice. J. Integrative Plant Biol. 51: 1126–1137.
Weir BS (1996) Genetic data analysis II: methods for discrete population genetic data. Sinauer, Sunderland, MA, pp. 445.
Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, and Holland JB (2005) A unified mixed-model method for association map** that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208.
Zong X, Redden RJ, Liu Q, Wang S, Guan J, Liu J, Xu Y, Liu X, Gu J, Yan L, Ades P, Ford R (2009) Analysis of a diverse global Pisum sp. collection and comparison to a Chinese local P. sativum collection with microsatellite markers. Theor. Appl. Genet. 18: 193–204.
Zong X-X, Guan J-P, Wang S-M, and Liu Q-C (2008) Genetic diversity among Chinese pea (Pisum sativum L.) landraces as revealed by SSR markers. Acta Agron. Sin. 34: 1330–1338.
Author information
Authors and Affiliations
Corresponding author
Additional information
S.-J. Kwon and A. F. Brown contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kwon, SJ., Brown, A.F., Hu, J. et al. Genetic diversity, population structure and genome-wide marker-trait association analysis emphasizing seed nutrients of the USDA pea (Pisum sativum L.) core collection. Genes Genom 34, 305–320 (2012). https://doi.org/10.1007/s13258-011-0213-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-011-0213-z