Abstract
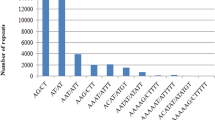
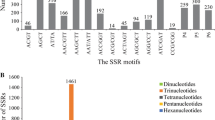
This study reports on the detection of additional expressed sequence tags (EST) derived simple sequence repeat (SSR) markers for the oil palm. A large collection of 19243 Elaeis guineensis ESTs were assembled to give 10258 unique sequences, of which 629 ESTs were found to contain 722 SSRs with a variety of motifs. Dinucleotide repeats formed the largest group (45.6%) consisting of 66.9% AG/CT, 21.9% AT/AT, 10.9% AC/GT and 0.3% CG/CG motifs. This was followed by trinucleotide repeats, which is the second most abundant repeat types (34.5%) consisting of AAG/CTT (23.3%), AGG/CCT (13.7%), CCG/CGG (11.2%), AAT/ATT (10.8%), AGC/GCT (10.0%), ACT/AGT (8.8%), ACG/CGT (7.6%), ACC/GGT (7.2%), AAC/GTT (3.6%) and AGT/ACT (3.6%) motifs. Primer pairs were designed for 405 unique EST-SSRs and 15 of these were used to genotype 105 E. guineensis and 30 E. oleifera accessions. Fourteen SSRs were polymorphic in at least one germplasm revealing a total of 101 alleles. The high percentage (78.0%) of alleles found to be specific for either E. guineensis or E. oleifera has increased the power for discriminating the two species. The estimates of genetic differentiation detected by EST-SSRs were compared to those reported previously. The transferability across palm taxa to two Cocos nucifera and six exotic palms is also presented. The polymerase chain reaction (PCR) products of three primer-pairs detected in E. guineensis, E. oleifera, C. nucifera and Jessinia bataua were cloned and sequenced. Sequence alignments showed mutations within the SSR site and the flanking regions. Phenetic analysis based on the sequence data revealed that C. nucifera is closer to oil palm compared to J. bataua; consistent with the taxanomic classification.
Similar content being viewed by others
References
Adin A., Weber J. C., Montes C. S., Vidaurre H., Vosman B. and Smulders M. J. M. 2004 Genetic differentiation and trade among populations of peach palm (Bactris gasipaes Kunth) in the Peruvian Amazon — implications for genetic resource management. Theor. Appl. Genet. 108, 1564–1573.
Billotte N., Risterucci A. M., Barcelos E., Noyer J. L., Amblard P. and Baurens F. C. 2001 Development, characterisation, and across-taxa utility of oil palm (Elaeis guineensis Jacq). microsatellite markers. Genome 44, 413–425.
Castillo A., Budak H., Varshney R. K., Dorado G., Graner A. and Hernandez P. 2008 Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol. 8, 97–105.
Cho J. C. and Tiedje J. M. 2001 Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66, 5448–5456.
Cordeiro G.M., Casu R., McLntyre C. L., Manners J.M. and Henry R. J. 2001 Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci. 160, 1115–1123.
Corley R. H. V. and Tinker P. B. 2003 The oil palm, 4th edition. Wiley-Blackwell, Oxford, UK.
Hayati A., Wickneswari R., Maiura I. and Rajanaidu N. 2004 Genetic diversity of oil palm (Elaeis guineensis Jacq.) germplasm collections from Africa: implications for improvement and conservation of genetic resources. Theor. Appl. Genet. 108, 1274–1284.
Jorgensen S., Hamrick J. L. and Well P. V. 2002 Regional patterns of genetic diversity in Pinus Flexilis (pinaceae) reveal complex species history. Am. J. Bot. 89, 792–800.
Low E. T. L., Halimah A., Boon S. H., Elyana M. S., Tan C. Y., Ooi L. C. L. et al. 2008 Oil palm (Elaeis guineensis Jacq.) tissue culture ESTs: identifying genes associated with callogenesis and embryogenesis. BMC Plant Biol. 8, 62.
Maizura I., Rajanaidu N., Zakri A. H. and Cheah S. C. 2006 Assesment of genetic diversity in oil palm (Elaeis guineensis Jacq.) using restriction fragment length polymorphism (RFLP). Genet. Res. Crop Evol. 53, 187–195.
Nei M. 1978 Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590.
Perera L., Russell J. R., Provan J. and Powell W. 2000 Use of microsatellite DNA markers to investigate the level of genetic diversity and population genetic structure of coconut (Cocos nucifera L.). Genome 43, 15–21.
Perera L., Russell J. R., Provan J. and Powell W. 2001 Level and distribution of genetic diversity of coconut (Cocos nucifera L., var. Typica form typica) from Sri Lanka assessed by microsatellite markers. Euphytica 122, 381–389.
Purba A. R., Noyer J. L., Baudouin L., Perrier X., Hamon S. and Lagoda P. J. L. 2000 A new aspect of genetic diversity of Indonesian oil palm (Elaeis guineensis Jacq.) revealed by isoenzyme and AFLP markers and its consequences for breeding. Theor. Appl. Genet. 101, 956–961.
Rajanaidu N., Jalani B. S., Kushairi A. and Rao V. 1999 Oil palm genetic resources-collection, evaluation, utilization and conser-vation. In Proceeding of the symposium on the science of oil palm breeding (ed. N. Rajanaidu and B. S. Jalani), pp. 219–255. PORIM, Bangi, Malaysia.
Rival A., Buele T., Barre P., Hamon S., Duval Y. and Noirot M. 1997 Comparative flow cytometric estimation of nuclear DNA content in oil palm (Elaeis guineensis Jacq.) tissue cultures and seed-derived plants. Plant Cell Rep. 16, 884–887.
Saitou N. and Nei M. 1987 The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sambanthamurthi R., Singh R., Parvee G. K. A., Ong-Abdullah M. and Kushairi 2009 Opportunities for the oil palm via breeding and biotechnology. In Breeding plantation tree crops (ed. S. M. Jain and P.M. Priyadarshan), pp. 377–421. Springer, New York, USA.
Singh R., Noorhariza M. Z., Ting N. C., Rozana R., Tan S. G., Low E. T. L. et al. 2008 Exploiting an oil palm EST database for the development of gene-derived and their exploitation for assessment of genetic diversity. Biologia 63, 227–235.
Swofford D. L. and Selander R. B. 1989 BIOSYS-1 A computer program for the analysis of allelic variation in population genetics and biochemical systematics. Release 1.7. Illinois Natural History Survey, Champaign, Illinois, USA.
Tang S. X., Okashah R. A., Cordonnier-Pratt M.-M., Pratt L. H., Johnson V. E., Taylor C. A. et al. 2009 EST and EST-SSR marker resources for Iris. BMC Plant Biol. 9, 72.
Thiel T., Michalek W., Varshney R. K. and Graner A. 2003 Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422.
Varshney R. K., Sorrells M. E. and Graner A. 2005 Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 23, 48–55.
Zehdi S., Trifi M., Billotte N., Merrakchi M. and Pintaud J. C. 2005 Genetic diversity of Tunisian date palms (Phoenix dactylifera L.) revealed by nuclear microsatellite polymorphism. Hereditas 141, 278–287.
Zehdi S., Sakka H., Rhouma A., Ould Mohamed Salem A., Marrakchi M. and Trifi M. 2004 Analysis of Tunisian date palm germplasm using simple sequence repeat primers. Afr. J. Biotechnol. 3, 215–219.
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ting, NC., Zaki, N.M., Rosli, R. et al. SSR mining in oil palm EST database: application in oil palm germplasm diversity studies. J Genet 89, 135–145 (2010). https://doi.org/10.1007/s12041-010-0053-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-010-0053-7