Abstract
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis (MTB) and considered as serious public health concern worldwide which kills approximately five thousand people every day. Therefore, TB drug development efforts are in gigantic need for identification of new potential chemical agents to eradicate TB from the society. The bacterial DNA gyrase B (GyrB) protein as an experimentally widely accepted effective drug target for the development of TB chemotherapeutics. In the present study, advanced pharmacoinformatics approaches were used to screen the Mcule database against the GyrB protein. Based on a number of chemometric parameters, five molecules were found to be crucial to inhibit the GyrB. A number of molecular binding interactions between the proposed inhibitors and important active site residues of GyrB were observed. The predicted drug-likeness properties of all molecules were indicated that compounds possess characteristics to be the drug-like candidates. The dynamic nature of each molecule was explored through the molecular dynamics (MD) simulation study. Various analyzing parameters from MD simulation trajectory have suggested rationality of the molecules to be potential GyrB inhibitor. Moreover, the binding free energy was calculated from the entire MD simulation trajectories highlighted greater binding free energy values for all newly identified compounds also substantiated the strong binding affection towards the GyrB in comparison to the novobiocin. Therefore, the proposed molecules might be considered as potential anti-TB chemical agents for future drug discovery purposes subjected to experimental validation.

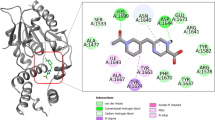
Graphical Abstract







Similar content being viewed by others
Availability of Data and Material
Not applicable.
References
Hoagland, D. T., Liu, J., Lee, R. B., & Lee, R. E. (2016). New agents for the treatment of drug-resistant mycobacterium tuberculosis. Advanced Drug Delivery Reviews, 102, 55–72.
Sotgiu, G., Centis, R., D'Ambrosio, L., & Migliori, G. B. (2015). Tuberculosis treatment and drug regimens. Cold Spring Harbor Perspectives in Medicine, 5(5), a017822.
Pourakbari, B., Mamishi, S., Mohammadzadeh, M., & Mahmoudi, S. (2016). First-line anti-tubercular drug resistance of mycobacterium tuberculosis in IRAN: a systematic review. Frontiers in Microbiology, 7, 1139.
Wu, X., Yang, J., Tan, G., Liu, H., Liu, Y., Guo, Y., Gao, R., Wan, B., & Yu, F. (2019). Drug resistance characteristics of mycobacterium tuberculosis isolates from patients with tuberculosis to 12 antituberculous drugs in China. Frontiers in Cellular and Infection Microbiology, 9, 1.
Dookie, N., Rambaran, S., Padayatchi, N., Mahomed, S., & Naidoo, K. (2018). Evolution of drug resistance in mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. Journal of Antimicrobial Chemotherapy, 73(5), 1138–1151.
Ghajavand, H., Kargarpour Kamakoli, M., Khanipour, S., Pourazar Dizaji, S., Masoumi, M., Rahimi Jamnani, F., Fateh, A., Yaseri, M., Siadat, S. D., & Vaziri, F. (2019). Scrutinizing the drug resistance mechanism of multi- and extensively-drug resistant mycobacterium tuberculosis: mutations versus efflux pumps. Antimicrobial Resistance & Infection Control, 8(1), 70.
Munir, A., Kumar, N., Ramalingam, S. B., Tamilzhalagan, S., Shanmugam, S. K., Palaniappan, A. N., Nair, D., Priyadarshini, P., Natarajan, M., Tripathy, S., Ranganathan, U. D., Peacock, S. J., Parkhill, J., Blundell, T. L., & Malhotra, S. (2019). Identification and characterization of genetic determinants of isoniazid and rifampicin resistance in mycobacterium tuberculosis in Southern India. Scientific Reports, 9(1), 10283.
Marimani, M., Ahmad, A., & Duse, A. (2018). The role of epigenetics, bacterial and host factors in progression of mycobacterium tuberculosis infection. Tuberculosis (Edinburgh, Scotland), 113, 200–214.
Jangam, C. S., Bhowmick, S., Chorge, R. D., Bharatrao, L. D., Patil, P. C., Chikhale, R. V., AlFaris, N. A., Altamimi, J. z., Wabaidur, S. M., & Islam, M. A. (2019). Pharmacoinformatics-based identification of anti-bacterial catalase-peroxidase enzyme inhibitors. Computational Biology and Chemistry, 83, 107136.
Bahuguna, A., & Rawat, D. S. (2020). An overview of new antitubercular drugs, drug candidates, and their targets. Medicinal Research Reviews, 40(1), 263–292.
Shirude, P. S., Madhavapeddi, P., Tucker, J. A., Murugan, K., Patil, V., Basavarajappa, H., Raichurkar, A. V., Humnabadkar, V., Hussein, S., Sharma, S., Ramya, V. K., Narayan, C. B., Balganesh, T. S., & Sambandamurthy, V. K. (2013). Aminopyrazinamides: novel and specific GyrB inhibitors that kill replicating and nonreplicating mycobacterium tuberculosis. ACS Chemical Biology, 8(3), 519–523.
Champoux, J. J. (2001). DNA topoisomerases: structure, function, and mechanism. Annual Review of Biochemistry, 70(1), 369–413.
Mdluli, K., & Ma, Z. (2007). Mycobacterium tuberculosis DNA gyrase as a target for drug discovery. Infectious Disorders Drug Targets, 7(2), 159–168.
van Doorn, H. R., An, D. D., de Jong, M. D., Lan, N. T., Hoa, D. V., Quy, H. T., Chau, N. V., Duy, P. M., Tho, D. Q., Chinh, N. T., Farrar, J. J., & Caws, M. (2008). Fluoroquinolone resistance detection in mycobacterium tuberculosis with locked nucleic acid probe real-time PCR. The International Journal of Tuberculosis and Lung Disease, 12(7), 736–742.
Aboul-Fadl, T., Abdel-Aziz, H. A., Abdel-Hamid, M. K., Elsaman, T., Thanassi, J., & Pucci, M. J. (2011). Schiff bases of indoline-2,3-dione: potential novel inhibitors of mycobacterium tuberculosis (Mtb) DNA gyrase. Molecules, 16(9), 7864–7879.
Brvar, M., Perdih, A., Hodnik, V., Renko, M., Anderluh, G., Jerala, R., & Solmajer, T. (2012). In silico discovery and biophysical evaluation of novel 5-(2-hydroxybenzylidene) rhodanine inhibitors of DNA gyrase B. Bioorganic & Medicinal Chemistry, 20(8), 2572–2580.
Brvar, M., Perdih, A., Oblak, M., Masic, L. P., & Solmajer, T. (2010). In silico discovery of 2-amino-4-(2,4-dihydroxyphenyl)thiazoles as novel inhibitors of DNA gyrase B. Bioorganic & Medicinal Chemistry Letters, 20(3), 958–962.
Azam, M. A., & Thathan, J. (2017). Pharmacophore generation, atom-based 3D-QSAR and molecular dynamics simulation analyses of pyridine-3-carboxamide-6-yl-urea analogues as potential gyrase B inhibitors. SAR and QSAR in Environmental Research, 28(4), 275–296.
Islam, M. A., & Pillay, T. S. (2017). Identification of promising DNA GyrB inhibitors for tuberculosis using pharmacophore-based virtual screening, molecular docking and molecular dynamics studies. Chemical Biology & Drug Design, 90(2), 282–296.
Islam, M. A., & Pillay, T. S. (2019). Identification of promising anti-DNA gyrase antibacterial compounds using de novo design, molecular docking and molecular dynamics studies. Journal of Biomolecular Structure & Dynamics, 1–12. https://doi.org/10.1080/07391102.2019.1617785.
Jeankumar, V. U., Renuka, J., Santosh, P., Soni, V., Sridevi, J. P., Suryadevara, P., Yogeeswari, P., & Sriram, D. (2013). Thiazole-aminopiperidine hybrid analogues: design and synthesis of novel mycobacterium tuberculosis GyrB inhibitors. European Journal of Medicinal Chemistry, 70, 143–153.
Kumar Muthyala, M. K., Jamullamudi, R. N., Sangeeta, G. P. V., & Kurre, P. N. (2018). Identification of N-Benzylated Indole Mannich bases as potential anti TB agents by using computational studies and molecular hybridization technique. Current Computer-Aided Drug Design, 14(3), 200–206.
Chopra, S., Matsuyama, K., Tran, T., Malerich, J. P., Wan, B., Franzblau, S. G., Lun, S., Guo, H., Maiga, M. C., Bishai, W. R., & Madrid, P. B. (2012). Evaluation of gyrase B as a drug target in mycobacterium tuberculosis. The Journal of Antimicrobial Chemotherapy, 67(2), 415–421.
Kamsri, P., Punkvang, A., Hannongbua, S., Suttisintong, K., Kittakoop, P., Spencer, J., Mulholland, A. J., & Pungpo, P. (2019). In silico study directed towards identification of the key structural features of GyrB inhibitors targeting MTB DNA gyrase: HQSAR, CoMSIA and molecular dynamics simulations. SAR and QSAR in Environmental Research, 30(11), 775–800.
Tari, L. W., Trzoss, M., Bensen, D. C., Li, X., Chen, Z., Lam, T., Zhang, J., Creighton, C. J., Cunningham, M. L., Kwan, B., Stidham, M., Shaw, K. J., Lightstone, F. C., Wong, S. E., Nguyen, T. B., Nix, J., & Finn, J. (2013). Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. Bioorganic & Medicinal Chemistry Letters, 23(5), 1529–1536.
Maharaj, Y., & Soliman, M. E. (2013). Identification of novel gyrase B inhibitors as potential anti-TB drugs: homology modelling, hybrid virtual screening and molecular dynamics simulations. Chemical Biology & Drug Design, 82(2), 205–215.
Saxena, S., Renuka, J., Yogeeswari, P., & Sriram, D. (2014). Discovery of novel mycobacterial DNA Gyrase B inhibitors: in silico and in vitro biological evaluation. Molecular Informatics, 33(9), 597–609.
Kale, M. G., Raichurkar, A., Hameed, P. S., Waterson, D., McKinney, D., Manjunatha, M. R., Kranthi, U., Koushik, K., Jena, L., Shinde, V., Rudrapatna, S., Barde, S., Humnabadkar, V., Madhavapeddi, P., Basavarajappa, H., Ghosh, A., Ramya, V. K., Guptha, S., Sharma, S., Vachaspati, P., Kumar, K. N., Giridhar, J., Reddy, J., Panduga, V., Ganguly, S., Ahuja, V., Gaonkar, S., Kumar, C. N., Ogg, D., Tucker, J. A., Boriack-Sjodin, P. A., de Sousa, S. M., Sambandamurthy, V. K., & Ghorpade, S. R. (2013). Thiazolopyridine ureas as novel antitubercular agents acting through inhibition of DNA Gyrase B. Journal of Medicinal Chemistry, 56(21), 8834–8848.
Barancokova, M., Kikelj, D., & Ilas, J. (2018). Recent progress in the discovery and development of DNA gyrase B inhibitors. Future Medicinal Chemistry, 10(10), 1207–1227.
Durcik, M., Tammela, P., Barancokova, M., Tomasic, T., Ilas, J., Kikelj, D., & Zidar, N. (2018). Synthesis and evaluation of N-Phenylpyrrolamides as DNA Gyrase B inhibitors. ChemMedChem, 13(2), 186–198.
Zidar, N., Macut, H., Tomasic, T., Peterlin Masic, L., Ilas, J., Zega, A., Tammela, P., & Kikelj, D. (2019). New N-phenyl-4,5-dibromopyrrolamides as DNA gyrase B inhibitors. Medchemcomm, 10(6), 1007–1017.
Durcik, M., Lovison, D., Skok, Z., Durante Cruz, C., Tammela, P., Tomasic, T., Benedetto Tiz, D., Draskovits, G., Nyerges, A., Pal, C., Ilas, J., Peterlin Masic, L., Kikelj, D., & Zidar, N. (2018). New N-phenylpyrrolamide DNA gyrase B inhibitors: optimization of efficacy and antibacterial activity. European Journal of Medicinal Chemistry, 154, 117–132.
Zhang, X., Perez-Sanchez, H., & Lightstone, F. C. (2017). A comprehensive docking and MM/GBSA rescoring study of ligand recognition upon binding antithrombin. Current Topics in Medicinal Chemistry, 17(14), 1631–1639.
Wang, C., Greene, D., **ao, L., Qi, R., & Luo, R. (2017). Recent developments and applications of the MMPBSA method. Frontiers in Molecular Biosciences, 4, 87.
Willems, A. R., Tahlan, K., Taguchi, T., Zhang, K., Lee, Z. Z., Ichinose, K., Junop, M. S., & Nodwell, J. R. (2008). Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. Journal of Molecular Biology, 376(5), 1377–1387.
(2018). Schrödinger Release 2018-4: Maestro, Schrödinger,, LLC, New York.
(2018). Schrödinger Release 2018-4: Prime, Schrödinger, LLC, New York, NY.
William, L., Jorgensen, D. S., & Maxwell, J. T. R. (1996). Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society, 118, 11225–11236.
Halgren, T. A., Murphy, R. B., Friesner, R. A., Beard, H. S., Frye, L. L., Pollard, W. T., & Banks, J. L. (2004). Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry, 47(7), 1750–1759.
Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., Sanschagrin, P. C., & Mainz, D. T. (2006). Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196.
Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461.
Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7(1), 42717.
Kumari, R., Kumar, R., Open Source Drug Discovery, C., & Lynn, A. (2014). g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962.
Islam, M. A., & Pillay, T. S. (2019). Beta-secretase inhibitors for Alzheimer’s disease: identification using pharmacoinformatics. Journal of Biomolecular Structure & Dynamics, 37(2), 503–522.
Taha, M. O., Habash, M., Al-Hadidi, Z., Al-Bakri, A., Younis, K., & Sisan, S. (2011). Docking-based comparative intermolecular contacts analysis as new 3-D QSAR concept for validating docking studies and in silico screening: NMT and GP inhibitors as case studies. Journal of Chemical Information and Modeling, 51(3), 647–669.
Wang, J., Zhao, C., Tu, J., Yang, H., Zhang, X., Lv, W., & Zhai, H. (2019). Design of novel quinoline-aminopiperidine derivatives as mycobacterium tuberculosis (MTB) GyrB inhibitors: an in silico study. Journal of Biomolecular Structure & Dynamics, 37(11), 2913–2925.
Acknowledgments
The authors would like to extend their sincere appreciation to the Researchers Supporting Project for funding this work through Research Number (RSP-2020/138), King Saud University, Riyadh, Saudi Arabia.
Funding
The authors would like to extend their sincere appreciation to the Researchers Supporting Project for funding this work through Research Number (RSP-2019/138), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Pranjali Mahadeo Tambe: data curation, investigation and methodology; Shovonlal Bhowmick: research conceptualization, investigation and methodology, supervison, writing—review and editing; Sushil K. Chaudhary: research conceptualization, writing—review and editing; Mohammad Rizwan Khan: writing—review and editing; Saikh M Wabaidur: writing—review and editing; Mohd. Muddassir: research conceptualization, writing—review and editing; Preeti Chunarkar Patil: research conceptualization, writing—review and editing; Md Ataul Islam: research conceptualization, investigation and methodology, supervison, writing—review and editing
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Code Availability
Not applicable.
Computational Resource
The CHPC (www.chpc.ac.za), Cape Town, South Africa is thankfully acknowledged for computational resources and tools.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 329 kb).
Rights and permissions
About this article
Cite this article
Tambe, P.M., Bhowmick, S., Chaudhary, S.K. et al. Structure-Based Screening of DNA GyraseB Inhibitors for Therapeutic Applications in Tuberculosis: a Pharmacoinformatics Study. Appl Biochem Biotechnol 192, 1107–1123 (2020). https://doi.org/10.1007/s12010-020-03374-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03374-y