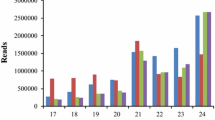
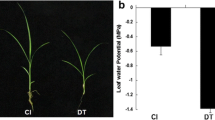
Abstract
Drought, in combination with high temperature and low humidity affects the productivity of Hevea brasiliensis, the natural rubber tree and its expansion to non-traditional regions. The genotypes of H. brasiliensis that perform well in traditional regions often failed in non-traditional regions thus necessitating breeding for stress-tolerant genotypes. This can be accomplished by adopting molecular-assisted selection method. Recent developments in identification of drought-responsive transcripts from H. brasiliensis and the findings on role of small RNAs indicate the possibility of employing them as markers for identification of suitable genotypes. In this study, we attempted to identify drought-responsive miRNAs from H. brasiliensis through next-generation sequencing (Illumina HiSeq) method. The results revealed the expression of 33 conserved and 32 novel drought-responsive miRNAs. Further, validation of differentially expressed miRNAs by quantitative expression analysis indicated the association of two novel miRNAs, viz., HbmiRn_63 and HbmiRn_42 and two conserved miRNAs, viz., miR168 and miR160 miRNAs with drought tolerance. These miRNAs can be employed as markers for drought tolerance after validation in a larger set of genotypes. This study opens up the possibility of employing miRNAs as markers for drought tolerance in Hevea.






Similar content being viewed by others
Abbreviations
- ARF:
-
Auxin response factors
- HMGR:
-
HMG-CoA reductase
- MFE:
-
Minimal folding free energy
- miRNA:
-
MicroRNA
- qPCR:
-
Quantitative PCR
- RRII:
-
Rubber Research Institute of India
References
Antolín-Llovera M, Leivar P, Arró M, Ferrer A, Boronat A, Campos N (2011) Modulation of plant HMG-CoA reductase by protein phosphatase 2A. Plant Signal Behav 6(8):1127–1131
Barozai MY, Baloch IA, Din M (2012) Identification of MicroRNAs and their targets in Helianthus. Mol Biol Rep 39:2523–2532
Barrera-Figueroa BE, Gao L, Diop NN, Wu Z, Ehlers JD, Roberts PA, Close TJ, Zhu J-K, Liu R (2011) Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol 11:127. https://doi.org/10.1186/1471-2229-11-127
Bonnet E, He Y, Billiau K, Peer YV (2010) TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26(12):1566–1568
Chandrashekar TR, Vijayakumar KR, George MJ, Sethuraj MR (1994) Response of few Hevea clones to partial irrigation during immature phase in a dry sub humid climatic region. Indian J Nat Rubber Res 7:114–119
Chinnusamy V, Zhu J, Zhou T, Zhu JK (2007) Small RNAs: big role in abiotic stress tolerance of plants. In: Jenks MA, Hasegawa PM, Jain SM (eds) Advances in molecular breeding toward drought and salt tolerant crops. Springer, New York, pp 223–260
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39(suppl_2):W155–W159
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432:231–235
Devakumar AS, Sathik MB, Jacob J, Annamalainathan K, Gawaiprakash P, Vijayakumar KR (1998) Effects of atmospheric and soil drought on growth and development of Hevea brasiliensis. J Rubber Res 1(3):190–198
Din M, Barozai MY, Baloch IA (2014) Identification and functional analysis of new conserved microRNAs and their targets in potato (Solanum tuberosum L.). Turk J Bot 38:1199–1213
Ding Y, Tao Y, Zhu C (2013) Emerging roles of microRNAs in the mediation of drought stress response in plants. J Exp Bot 64(11):3077–3086. https://doi.org/10.1093/jxb/ert164
Eldem V, Akçay UC, Ozhuner E, Bakır Y, Uranbey S, Unver T (2012) Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS One 7(12):e50298. https://doi.org/10.1371/journal.pone.0050298
Ferdous J, Hussain SS, Shi B-J (2015) Role of microRNAs in plant drought tolerance. Plant Biotechnol J 13:293–305
Ferdous J, Whitford R, Nguyen M, Brien C, Langridge P, Tricker PJ (2016) Drought-inducible expression of Hv-miR827 enhances drought tolerance in transgenic barley. Funct Integr Genom 17(2–3):279–292
Frazier TP, Sun G, Burklew CE, Zhang B (2011) Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol Biotechnol 49(2):159–165
Fu D, Ma B, Mason AS, ** technique in Brassica species. Plant Breed 132(4):375–381
Gebelin V, Argout X, Engchuan W, Pitollat B, Duan C, Montoro P, Leclercq J (2012) Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biol 12:18
Gebelin V, Leclercq J, Argout X, Chaidamsari T, Hu S, Tang C, Sarah G, Yang M, Montoro P (2013a) The small RNA profile in latex from Hevea brasiliensis trees is affected by tap** panel dryness. Tree Physiol 31:1084–1098
Gebelin V, Leclercq J, Chaorong T, Songnian H, Tang C, Montoro P (2013b) Regulation of MIR genes in response to abiotic stress in Hevea brasiliensis. Int. J Mol Sci 14:19587–19604
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240
Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5:3050
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460. https://doi.org/10.1016/j.pbi.2007.08.014
Iyer LM, Koonin EV, Aravind L (2004) Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle 3:1440–1450
Jaleel CA, Manivannan P, Wahid A, Farooq M, Somasundaram R, Panneerselvam R (2009) Drought stress in plants: a review on morphological characteristics and pigments composition. Int J Agric Biol 11:100–105
Kantar M, Unver T, Budak H (2010) Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genom 10:493–507
Kantar M, Lucas S, Budak H (2011) miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233:471–484
Krishan B (2017) Assessment of drought tolerance in few clones of natural rubber (Hevea brasiliensis) under dry hot climate of Odisha, India. J Exp Biol Agric Sci. https://doi.org/10.18006/2017.5(1).106.110
Kuruvilla L, Sathik MBM, Thomas M, Luke LP, Sumesh KV, Annamalainathan K (2016) Expression of miRNAs of Hevea brasiliensis under drought stress is altered in genotypes with varying levels of drought tolerance. Indian J Biotechnol 15:153–160
Kuruvilla L, Sathik MBM, Thomas M, Luke LP, Sumesh KV (2017)) Identification and validation of cold responsive microRNAs of Hevea brasiliensis using high throughput sequencing. J Crop Sci Biotechnol 20(5):369–377
Lawlor DW (2013) Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. J Exp Bot 64:83–108
Leclercq J, Martin F, Sanier C, Clement-Vidal A, Fabre D, Oliver G, Lardet L, Ayar A, Peyramard M, Montoro P (2012) Over-expression of a cytosolic isoform of the HbCuZnSOD gene in Hevea brasiliensis changes its response to a water deficit. Plant Mol Biol 80:255e272
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Lertpanyasampatha M, Gao L, Kongsawadworakul P, Viboonjum U, Chrestin H, Liu R, Chen X, Narangaajavana J (2012) Genome-wide analysis of microRNAs in rubber tree (Hevea brasiliensis L.) using high-throughput sequencing. Planta 236(2):437–445
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, ** H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post transcriptionally to promote drought resistance. Plant Cell 20:2238–2251. https://doi.org/10.1105/tpc.108.059444
Li B, Yin W, **a X (2009) Identification of microRNAs and their targets from Populus euphratica. Biochem Biophys Res Commun 388:272–277
Li W, Wang T, Zhang Y, Li Y (2015) Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J Exp Bot. https://doi.org/10.1093/jxb/erv450
Li W, Wang T, Zhang Y, Li Y (2016) Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J Exp Bot 67:175–194. https://doi.org/10.1093/jxb/erv450
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Liu W, Yu W, Hou L, Wang X, Zheng F, Wang W, Liang D, Yang H, ** Y (2014) Analysis of miRNAs and their targets during adventitious shoot organogenesis of Acacia crassicarpa. PLoS One 9(4):e93438. https://doi.org/10.1371/journal.pone.0093438
Liu M, Yu H, Zhao G, Huang Q, Lu Y, Ouyang B (2017) Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genom. https://doi.org/10.1186/s12864-017-3869-1
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408
Luke LP, Sathik MBM, Thomas M, Kuruvilla L, Sumesh KV, Annamalainathan K (2015) Quantitative expression analysis of drought responsive genes in genotypes of Hevea with varying levels of drought tolerance. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-015-0288-0
Mohanakrishna T, Bhasker CVS, Rao SP, Chandrashaker TR, Sethuraj MR, Vijayakumar KR (1991) Effect of irrigation on physiological performance of immature plants of Hevea brasiliensis in North Konkan. Indian J Nat Rubber Res 4:36–45
Mudgil Y, Shiu S-H, Stone SL, Salt JN, Goring DR (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-Box E3 ubiquitin ligase family. Plant Physiol 134(1):59–66. https://doi.org/10.1104/pp.103.029553
Ni Z, Hu Z, Jiang Q, Zhang H (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol 82:113–129. https://doi.org/10.1007/s11103-013-0040-5
Noman A, Aqeel M (2017) miRNA-based heavy metal homeostasis and plant growth. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-017-8593-5
Noman A, Fahad S, Aqeel M, Ali U, Amanullah, Anwar S, Baloch SK, Zainab M (2017) miRNAs: major modulators for crop growth and development under abiotic stresses. Biotechnol Lett 39(5):685–700. https://doi.org/10.1007/s10529-017-2302-9
Razna K, Hlavackova L, Bezo M, Ziarovska J, Haban M, Slukova Z, Pernisova M (2015) Application of the RAPD and miRNA markers in the genoty** of Silybum marianum (L.) Gaertn. Acta phytotechnica et zootechnica 18(4):83–89. https://doi.org/10.15414/afz.2015.18.04.83-89
Sathik MBM, Luke LP, Kuruvilla L, Thomas M (2018) De novo transcriptome analysis of abiotic stress responsive genes of Hevea brasiliensis. Mol Breed. https://doi.org/10.1007/s11032-018-0782-5
Sethuraj MR, Rao CG, Raghavendra AS (1984) The pattern of latex flow from rubber tree (Hevea brasiliensis) in relation to water stress. J Cell Biochem Suppl 8B:236
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6(5):410–417
Shriram V, Kumar V, Devarumath RM, Khare TS, Wani SH (2016) MicroRNAs as potential targets for abiotic stress tolerance in plants. Front Plant Sci 7:817. https://doi.org/10.3389/fpls.2016.00817
Shuai P, Liang D, Zhang Z, Yin W, **a X (2013) Identification of drought-responsive and novel Populus trichocarpa microRNAs by high throughput sequencing and their targets using degradome analysis. BMC Genom 14:233. https://doi.org/10.1186/1471-2164-14-233
Solofoharivelo MC, Walt AP, Stephan D, Burger JT, Murray SL (2014) MicroRNAs in fruit trees: discovery, diversity and future research directions. Plant Biol 16:856–865
Song JB, Gao S, Wang Y, Li BW, Zhang YL, Yang ZM (2016) miR394 and its target gene LCR are involved in cold stress response in Arabidopsis. Plant Gene 5:56–64. https://doi.org/10.1016/j.plgene.2015.12.001
Sreelatha S, Simon SP, Kurup GM, Vijayakumar KR (2007) Biochemical mechanisms associated with low yield during stress in Hevea genotype RRII 105. J Rubber Res 10:107–150
Sreelatha S, Mydin KK, Simon SP, Krishnakumar R, Jacob J, Annamalainathan K (2011) Seasonal variations in yield and associated biochemical changes in RRII 400 series genotypes of Hevea brasiliensis. Nat Rubber Res 24:117–123
Sumesh KV, Satheesh PR, Annamalainathan K, Krishnakumar R, Thomas M, Jacob J (2011) Physiological evaluation of a few modern Hevea clones for intrinsic drought tolerance. Nat Rubber Res 24(1):61–67
Sunkar R, Li Y, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Cell 17:196–203
Tang S, Wang Y, Li Z, Gui Y, ** responsive small RNAs in tobacco (Nicotiana tabacum). BMC Plant Biol 12:28
Thomas M, Sathik MBM, Luke LP et al (2012a) Stress responsive transcripts and their association with drought tolerance in Hevea brasiliensis. J Plant Crops 40(3):180–187
Thomas M, Sathik MBM, Luke LP, Sumesh KV, Satheesh PR, Annamalainathan K, Jacob J (2012b) Screening of Drought Responsive transcripts of Hevea brasiliensis and Identification of Candidate Genes for Drought Tolerance. J Plant Biol 38 & 39:111–118
Thomas M, Sumesh KV, Sreelatha S et al (2014) Biochemical evaluation of RRII 400 series clones of Hevea brasiliensis for drought tolerance. Indian J Agric Biochem 27(1):35–39
Thomas M, Xavier SM, Sumesh KV, Annamalainathan K, Nair DB, Mercy MA (2015) Identification of potential drought tolerant Hevea germplasm accessions using physiological and biochemical parameters. Rubber Sci 28(1):62–69
Vaucheret H (2008) Plant argonautes. Trends Plant Sci 13(7):350–358
Voinnet O (2009) Origin, biogenesis and activity of plant microRNAs. Cell 136:669–687
Wang X, Gui S, Pan L, Hu J, Ding Y (2016) Development and characterization of polymorphic microRNA-based microsatellite markers in Nelumbo nucifera(Nelumbonaceae). Appl Plant Sci 4(1). https://doi.org/10.3732/apps.1500091
**a R, Zhu H, An Y-q, Beers EP, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13(6):R47
Yadav CB, Muthamilarasan M, Pandey G, Khan Y (2014) Development of novel microRNA-based genetic markers in foxtail millet for genoty** applications in related grass species. Mol Breed 34(4):2219–2224
Zhang B (2015) MicroRNAs: a new target for improving plant tolerance to abiotic stress. J Exp Bot 66(7):1749–1761. https://doi.org/10.1093/jxb/erv013
Zhang B, Wang Q (2015) MicroRNA-based biotechnology for plant improvement. J Cell Physiol 230:1–15. https://doi.org/10.1002/jcp.24685
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61(15):4157–4168. https://doi.org/10.1093/jxb/erq237
Zhu Q-W, Luo Y-P (2013) Identification of miRNAs and their targets in tea (Camellia sinensis). J Zhejiang Univ Sci B 14(10):916–923
Acknowledgements
The authors wish to thank Dr. K. Annamalainathan (Joint Director, RRII), Dr. Kavitha K. Mydin (Joint Director, RRII) and Dr. James Jacob (Director of Research, RRII) for their constant support and encouragement throughout the course of the work. Authors also wish to thank Dr. Shammi Raj (Principal Scientist, meteorology) and Dr. K.V Sumesh (Scientist, Plant Physiology) for their help in data analysis and Ms. Smitha M Xavier (Research Scholar) for providing germplasm materials. The authors also thank Dr. Deepthy Antony (Senior Scientist, Plant Breeding) for critical reading and correcting the manuscript. Ms. Linu Kuruvilla is grateful to Council of Scientific and Industrial Research, New Delhi for the Senior Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Wojtaszek.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kuruvilla, L., Sathik, M., Luke, L.P. et al. Identification and validation of drought-responsive microRNAs from Hevea brasiliensis. Acta Physiol Plant 41, 14 (2019). https://doi.org/10.1007/s11738-018-2803-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-018-2803-8