Abstract
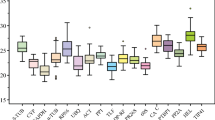
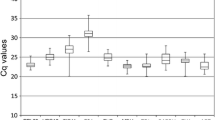
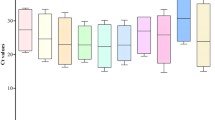
RT-qPCR is a widely used method in gene expression and transcriptome studies. Normalization based on reference genes is necessary to accurately analyze RT-qPCR data. Thus, an accurate and systematic evaluation of these reference genes before experiments are conducted is necessary. In this study, 18 candidate reference genes were evaluated under various experimental conditions covering a range of tissue types and cultivars, NaCl, CaCl2 and temperature treatments, hormones (6-BA, ABA and NAA) and a set of osmotic stress (mannitol and PEG6000) treatments. Gene expression across 48 pear samples was evaluated using geNorm, NormFinder and BestKeeper statistical algorithms. Actin2/7 (ACT2/7), ubiquitin extension protein (UBI) and Yellow-leaf-specific gene 8 (YLS8) exhibited the most stable expression across all the pear samples tested. While in the other experimental groups, different sets of samples had their own best reference genes. In addition, the gene expression of PbCBL7, a member of the calcineurin B-like protein, was measured across all the 48 samples using the best three reference genes, it displayed variation in gene expression across different tissues and cultivars, and exhibited diverse up- or down-regulated expression patterns under various treatments, which indicate that PbCBL7 may play a role in response to specific abiotic stress in pear. These results are valuable for future research on gene expression and abiotic stress tolerance in pear.






Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- bp:
-
Base pair
- NAA:
-
1-Naphthaleneacetic acid
- PEG6000:
-
Polyethylene Glycol 6000
- RT-qPCR:
-
Quantitative real-time polymerase chain reaction
- 6-BA:
-
6-Benzylaminopurine
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14
Chang E, Shi S, Liu J, Cheng T, Xue L, Yang X, Yang W, Lan Q, Jiang Z (2012a) Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS One 7:e33278
Chang YH, Li H, Cong Y, Lin J, Sheng BL (2012b) Characterization and expression of a phytochelatin synthase gene in birch-leaf pear (Pyrus betulaefolia Bunge). Plant Mol Biol Rep 30:1329–1337
Chi XY, Hu RB, Yang QL, Zhang XW, Pan LJ, Chen N, Chen MN (2012) Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genomics 287:167–176
Coito JL, Rocheta M, Carvalho L, Amâncio S (2012) Microarray-based uncovering reference genes for quantitative real time PCR in grapevine under abiotic stress. BMC Res Notes 5:220
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53:28–37
Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Garson JA, Grant PR, Ayliffe U, Ferns RB, Tedder RS (2005) Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J Virol Methods 126:207–213
Gonzalez-Verdejo CL, Die JV, Nadal S, Jimenez-Marin A, Moreno MT, Roman B (2008) Selection of housekee** genes for normalization by real-time RT-PCR: analysis of Or-MYB1 gene expression in Orobanche ramosa development. Anal Biochem 379:176–181
Gu Z, Ma B, Jiang Y, Chen Z, Su X, Zhang H (2008) Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene 415:1–12
Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Han B, Yang Z, Samma MK, Wang R, Shen W (2013) Systematic validation of candidate reference genes for qRT-PCR normalization under iron deficiency in Arabidopsis. Biometals 26:403–413
Hu RB, Fan CM, Li HY, Zhang QZ, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284
Huis R, Hawkins S, Neutelings G (2010) Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol 10:71
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekee** genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B (2005) Enhancing Arabidopsis salt and drought stress tolerance by Chemical priming for its abscisic acid responses. Plant Physiol 139:267–274
Jarošová J, Kundu J (2010) Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol 10:146
Jian B, Liu B, Bi YR, Hou WS, Wu CX, Han TF (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Jiang Y, Liang G, Yu D (2012) Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant 5:1375–1388
Klie M, Debener T (2011) Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res Notes 4:518
Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96:4718–4723
Li QF, Sun SSM, Yuan DY, Yu HX, Gu MH, Liu QQ (2010) Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep 28:49–57
Løvdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization intomato subjected to nitrogen, cold, and light stress. Anal Biochem 387:238–242
Luan S (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14:37–42
Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, Boava LP, Rodrigues CM, Machado MA (2012) Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One 7:e31263
Marino ER, Borges AA, Perez AB, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Maroufi A, Bockstaele EV, Loose MD (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15
Martin RC, Hollenbeck VG, Dombrowski JE (2008) Evaluation of reference genes for quantitative RT-PCR in Lolium perenne. Crop Sci 48:1881–1887
Marum L, Miguel A, Ricardo CP, Miguel C (2012) Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS One 7:e35113
McDowell JM, Huang S, McKinney EC, An YQ, Meagher RB (1996) Structure and evolution of the actin gene family in Arabidopsis thaliana. Genet 142:587–602
Morgante CV, Guimarães PM, Martins ACQ, Araújo ACG, Leal-Bertioli SCM, Bertioli DJ, Brasileiro ACM (2011) Reference genes for quantitative reverse transcription-polymerase chain reaction expression studies in wild and cultivated peanut. BMC Res Notes 4:339
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekee** gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Obrero Á, Die JV, Romén B, Gómez P, Nadal S, Gonzalez-Verdejo CL (2011) Selection of reference genes for gene expression studies in zucchini (Cucurbita pepo) using qPCR. J Agric Food Chem 59:5402–5411
Pandey GK, Cheong YH, Kim KN, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16:1912–1924
Pandey S, Nookaraju A, Upadhyaya CP, Gururani MA, Venkatesh J, Kim DH, Park SW (2011) An update on biotechnological approaches for improving abiotic stress tolerance in tomato. Crop Sci 51:2303–2324
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekee** genes, differentially regulated target genes, and sample integrity: bestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349
Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283:233–241
Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11:2393–2406
Shi J, Liu M, Shi J, Zheng G, Wang Y, Wang J, Chen Y, Lu C, Yin W (2012) Reference gene selection for qPCR in Ammopiptanthus mongolicus under abiotic stresses and expression analysis of seven ROS-scavenging enzyme genes. Plant Cell Rep 31:1245–1254
Silveira ÉD, Alves-Ferreira M, Guimarães LA, da Silva FR, Carneiro VT (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol 9:84
Tong ZG, Gao ZH, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10:71
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Vashisth T, Johnson LK, Malladi A (2011) An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep 30:2167–2176
Visioni A, Tondelli A, Francia E, Pswarayi A, Malosetti M, Russell J, Thomas W, Waugh R, Pecchioni N, Romagosa I, Comadran J (2013) Genome-wide association map** of frost tolerance in barley (Hordeum vulgare L.). Plant. Genomics 14:424
Wan HJ, Zhao ZG, Qian CT, Sui YH, Malik AA, Chen JF (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Wan H, Yuan W, Ruan M, Ye Q, Wang R, Li Z, Zhou G, Yao Z, Zhao J, Liu S, Yang Y (2011) Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem Biophys Res Commun 416:24–30
Wu J, Wang Z, Shi Z (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Xu Y, Zhu X, Gong Y, Xu L, Wang Y, Liu L (2012) Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem Biophys Res Commun 424:398–403
Yang Y, Hou S, Cui G, Chen S, Wei J, Huang L (2010) Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol Biol Rep 37:507–513
Zhang H, Yin W, **a X (2008) Calcineurin B-Like family in Populus: comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul 56:129–140
Zhang X, Allan AC, Yi Q, Chen L, Li K, Shu Q, Su J (2011) Differential Gene expression analysis of Yunnan red pear, Pyrus Pyrifolia, during fruit skin coloration. Plant Mol Biol Rep 29:305–314
Zhang H, Zhang S, Qin G, Wang L, Wu T, Qi K, Zhang S (2013) Molecular cloning and expression analysis of a gene for sucrose transporter from pear (Pyrus bretschneideri Rehd.) fruit. Plant Physiol Biochem 73:63–69
Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, Chen JY, Lu WJ, Li JG (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative realtime PCR in litchi under different experimental conditions. Plant Cell Rep 30:641–653
Zhu J, Zhang L, Li W, Han S, Yang W, Qi L (2013) Reference gene selection for quantitative real-time PCR normalization in caragana intermedia under different abiotic stress conditions. PLoS One 8:e53196
Acknowledgments
This work was supported by grants from the Program for the National Natural Science Foundation of China (Grant No. 31372051) and Post-doctoral Foundation of Jiangsu, China (Grant No. 1302006B).
Conflict of interest
All authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by S. Abe.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, Y., Li, H., Li, X. et al. Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiol Plant 37, 40 (2015). https://doi.org/10.1007/s11738-015-1784-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1784-0