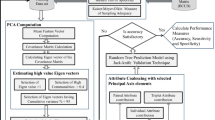
Abstract
Breast cancer is a widespread disease and one of the primary causes of cancer mortality among women all over the world. Computer-aided methods are used to assist medical doctors to make early diagnosis of the disease. The aim of this study is to build an effective prediction model for breast cancer diagnosis based on anthropometric data and parameters collected through routine blood analysis. The proposed approach innovatively exploits principal component analysis (PCA) technique cascaded by median filtering so as to transform original features into a form of containing less distractive noise not to cause overfitting. Since a generalized regression neural network (GRNN) model is adopted to classify patterns of the transformed features, the computational load imposed in the training of artificial neural network model is kept minimized thanks to the non-iterative nature of GRNN training. The proposed method has been devised and tested on the recent Breast Cancer Coimbra Dataset (BCCD) that contains 9 clinical features measured for each of 116 subjects. Outperforming all of the existing studies on BCCD, our method achieved a mean accuracy rate of 0.9773. Experimental results evidence that this study achieves the best prediction performance ever reported on this dataset. The fact that our proposed approach has accomplished such a boosted performance of breast cancer diagnosis based on routine blood analysis features offers a great potential to be used in a widespread manner to detect the disease in its inception phase.

Graphical abstract














Similar content being viewed by others
References
International Agency for Research on Cancer. https://www.iarc.fr/. Accessed 15 Jan 2019
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
World Health Organization. https://www.who.int/. Accessed 11 January 2019
New Global Cancer Data: GLOBOCAN, 2018. https://www.uicc.org/new-global-cancer-data-globocan-2018.
American Cancer Society, Cancer Facts & Figures 2019 Report. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2019.html.
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics. Ca-Cancer J Clin 69:7–34. https://doi.org/10.3322/caac.21551
Eyupoglu C (2018) Breast cancer classification using k-nearest neighbors algorithm. Online J Sci Technol 8(3):29–34
Jeleń Ł, Krzyżak A, Fevens T, Jeleń M (2016) Influence of feature set reduction on breast cancer malignancy classification of fine needle aspiration biopsies. Comput Biol Med 79:80–91. https://doi.org/10.1016/j.compbiomed.2016.10.007
Wolberg WH, Street WN, Mangasarian OL (1995) Breast Cancer Wisconsin Data Set, UCI Machine Learning Repository. http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/. Accessed 3 January 2019
Abdar M, Zomorodi-Moghadam M, Zhou X, Gururajan R, Tao X, Barua PD, Gururajan R (2018) A new nested ensemble technique for automated diagnosis of breast cancer. Pattern Recogn Lett (In Press) 2018. https://doi.org/10.1016/j.patrec.2018.11.004
Wang H, Zheng B, Yoon SW, Ko HS (2018) A support vector machine-based ensemble algorithm for breast cancer diagnosis. Eur J Oper Res 267(2):687–699. https://doi.org/10.1016/j.ejor.2017.12.001
Liu N, Qi ES, Xu M, Gao B, Liu GQ (2019) A novel intelligent classification model for breast cancer diagnosis. Comm Com Inf Sc 56(3):609–623. https://doi.org/10.1016/j.ipm.2018.10.014
Jafari-Marandi R, Davarzani S, Gharibdousti MS, Smith BK (2018) An optimum ANN-based breast cancer diagnosis: bridging gaps between ANN learning and decision-making goals. Appl Soft Comput 72:108–120. https://doi.org/10.1016/j.asoc.2018.07.060
Sheikhpour R, Sarram MA, Sheikhpour R (2016) Particle swarm optimization for bandwidth determination and feature selection of kernel density estimation based classifiers in diagnosis of breast cancer. Appl Soft Comput 40:113–131. https://doi.org/10.1016/j.asoc.2015.10.005
Peng L, Chen W, Zhou W, Li F, Yang J, Zhang J (2016) An immune-inspired semi-supervised algorithm for breast cancer diagnosis. Comput Methods Prog Biomed 134:259–265. https://doi.org/10.1016/j.cmpb.2016.07.020
Aličković E, Subasi A (2017) Breast cancer diagnosis using GA feature selection and rotation forest. Neural Comput & Applic 28(4):753–763. https://doi.org/10.1007/s00521-015-2103-9
Dora L, Agrawal S, Panda R, Abraham A (2017) Optimal breast cancer classification using Gauss–Newton representation based algorithm. Expert Syst Appl 85:134–145. https://doi.org/10.1016/j.eswa.2017.05.035
Nilashi M, Ibrahim O, Ahmadi H, Shahmoradi L (2017) A knowledge-based system for breast cancer classification using fuzzy logic method. Telematics Inform 34(4):133–144. https://doi.org/10.1016/j.tele.2017.01.007
Karabatak M (2015) A new classifier for breast cancer detection based on Naïve Bayesian. Measurement 72:32–36. https://doi.org/10.1016/j.measurement.2015.04.028
Shirazi AZ, Chabok SJSM, Mohammadi Z (2018) A novel and reliable computational intelligence system for breast cancer detection. Med Biol Eng Comput 56(5):721–732. https://doi.org/10.1007/s11517-017-1721-z
Patrício M, Pereira J, Crisóstomo J, Matafome P, Gomes M, Seiça R, Caramelo F (2018) Breast Cancer Coimbra Data Set, UCI Machine Learning Repository. https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Coimbra.
Li Y, Chen Z (2018) Performance evaluation of machine learning methods for breast cancer prediction. Appl Comput Math 7(4):212–216. https://doi.org/10.11648/j.acm.20180704.15
Livieris I, Pintelas E, Kanavos A, Pintelas P (2018) An improved self-labeled algorithm for cancer prediction. In: Cohen IR, Lajtha A, Lambris JD, Paoletti R, Rezaei N (eds) Advances in experimental medicine and biology. Publisher, Springer, pp 1–10
Aslan MF, Celik Y, Sabanci K, Durdu A (2018) Breast cancer diagnosis by different machine learning methods using blood analysis data. Int J Intelli Syst Appl Eng 6(4):289–293. https://doi.org/10.18201/ijisae.2018648455
Polat K, Sentürk U (2018) A novel ML approach to prediction of breast cancer: combining of mad normalization, KMC based feature weighting and AdaBoostM1 classifier. In: 2nd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), 2018, pp 1-4
Silva Araújo VJ, Guimarães AJ, de Campos Souza PV, Silva Rezende T, Souza Araújo V (2019) Using resistin, glucose, age and bmi and pruning fuzzy neural network for the construction of expert systems in the prediction of breast cancer. Mach Learn Knowl Extr 1(1):466–482. https://doi.org/10.3390/make1010028
Akben SB (2019) Determination of the blood, hormone and obesity value ranges that indicate the breast cancer, using data mining based expert system. IRBM 40(6):355–360. https://doi.org/10.1016/j.irbm.2019.05.007
Singh BK (2019) Determining relevant biomarkers for prediction of breast cancer using anthropometric and clinical features: a comparative investigation in machine learning paradigm. Biocybern Biomed Eng 39(2):393–409. https://doi.org/10.1016/j.bbe.2019.03.001
Dalwinder S, Birmohan S, Manpreet K (2019) Simultaneous feature weighting and parameter determination of neural networks using ant lion optimization for the classification of breast cancer. Biocybern Biomed Eng 40(1):337–351. https://doi.org/10.1016/j.bbe.2019.12.004
Ontiveros-Robles E, Melin P (2020) Toward a development of general type-2 fuzzy classifiers applied in diagnosis problems through embedded type-1 fuzzy classifiers. Soft Comput 24(1):83–99. https://doi.org/10.1007/s00500-019-04157-2
Abdel-Basset M, El-Shahat D, El-henawy I, de Albuquerque VHC, Mirjalili S (2020) A new fusion of grey wolf optimizer algorithm with a two-phase mutation for feature selection. Expert Syst Appl 139:112824. https://doi.org/10.1016/j.eswa.2019.112824
Patrício M, Pereira J, Crisóstomo J, Matafome P, Gomes M, Seiça R, Caramelo F (2018) Using Resistin, glucose, age and BMI to predict the presence of breast cancer. BMC Cancer 18(1):29. https://doi.org/10.1186/s12885-017-3877-1
Tukey JW (1977) Exploratory data analysis. Addison-Wesley, Reading, Mass
MathWorks Statistics and Machine Learning Toolbox. The MathWorks Inc., 2018
Salo F, Nassif AB, Essex A (2019) Dimensionality reduction with IG-PCA and ensemble classifier for network intrusion detection. Comput Netw 148:164–175. https://doi.org/10.1016/j.comnet.2018.11.010
Jackson JE (2005) A user’s guide to principal components. John Wiley & Sons
Justusson BI (1981) Median filtering: statistical properties. In: Two-dimensional digital signal processing. Springer, Berlin, Heidelberg, pp 161–196
Broesch JD (2008) Digital signal processing: instant access. Elsevier
Chen S, Cowan CF, Grant PM (1991) Orthogonal least squares learning algorithm for radial basis function networks. IEEE T Neural Networ 2(2):302–309. https://doi.org/10.1109/72.80341
Demuth HB, Beale MH, Hagan MT (2006) Neural network toolbox user’s guide. The MathWorks Inc
Powell MJD (1987) Radial basis functions for multivariable interpolation: a review. In: Mason JC, Cox MG (eds) Algorithms for approximation, Publisher: Clarendon Press. Imprint of Oxford University Press, New York, pp 143–167
Lowe D, Broomhead D (1988) Multivariable functional interpolation and adaptive networks. Nonl Phen Compl Syst 2(3):321–355
Hagan MT, Demuth HB, Beale MH, De Jesús O (1996) Neural network design, vol 20. Pws Pub, Boston
Bauer MM (1995) General regression neural network for technical use. University of Wisconsin-Madison, Master’s thesis
Hannan SA, Manza RR, Ramteke RJ (2010) Generalized regression neural network and radial basis function for heart disease diagnosis. Int J Comput Appl 7(13):7–13. https://doi.org/10.5120/1325-1799
Yavuz E, Kasapbaşı MC, Eyüpoğlu C, Yazıcı R (2018) An epileptic seizure detection system based on cepstral analysis and generalized regression neural network. Biocybern Biomed Eng 38(2):201–216. https://doi.org/10.1016/j.bbe.2018.01.002
Yavuz E, Eyupoglu C, Sanver U, Yazici R (2017) An ensemble of neural networks for breast cancer diagnosis. In: 2017 IEEE International Conference on Computer Science and Engineering (UBMK), pp 538-543
Yavuz E, Eyupoglu C (2019) A cepstrum analysis-based classification method for hand movement surface EMG signals. Med Biol Eng Comput 57(10):2179–2201. https://doi.org/10.1007/s11517-019-02024-8
Sun X, Liu J, Zhu K, Hu J, Jiang X, Liu Y (2019) Generalized regression neural network association with terahertz spectroscopy for quantitative analysis of benzoic acid additive in wheat flour. R Soc Open Sci 6(7):190485. https://doi.org/10.1098/rsos.190485
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45(4):427–437. https://doi.org/10.1016/j.ipm.2009.03.002
Witten I, Frank E, Hall M, Pal C (2017) Data mining: practical machine learning tools and techniques, 4th edn. Kaufmann, Morgan
Freund Y, Schapire RE (1996) Experiments with a new boosting algorithm. In: 13th International Conference on Machine Learning, pp 148–156. July, Bari, pp 3–6
Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK (2001) Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Comput 13(3):637–649. https://doi.org/10.1162/089976601300014493
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/A:1010933404324
Deep Learning for Java, Deeplearning4j. https://deeplearning4j.org/. Accessed 26 January 2020
John GH, Langley P (1995) Estimating continuous distributions in Bayesian classifiers. In: 10th Conference on Uncertainty in Artificial Intelligence (UAI’95), pp 338–345. August, Montréal, pp 18–20
Le Cessie S, Van Houwelingen JC (1992) Ridge estimators in logistic regression. J Roy Stat Soc C-App 41(1):191–201. https://doi.org/10.2307/2347628
Genkin A, Lewis DD, Madigan D (2004) Large-scale Bayesian logistic regression for text categorization. Technometrics 49(3):291–304. https://doi.org/10.1198/004017007000000245
Aha DW, Kibler D, Albert MK (1991) Instance-based learning algorithms. Mach Learn 6(1):37–66. https://doi.org/10.1007/BF00153759
Cleary JG, Trigg LE (1995) K*: an instance-based learner using an entropic distance measure. In: 12th international conference on machine learning, pp 108–114. July, Tahoe City, California, pp 9–12
Quinlan R (1993) C4.5: programs for machine learning. Morgan Kaufmann Publishers, San Mateo, CA
Landwehr N, Hall M, Frank E (2005) Logistic model trees. Mach Learn 59(1–2):161–205. https://doi.org/10.1007/s10994-005-0466-3
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yavuz, E., Eyupoglu, C. An effective approach for breast cancer diagnosis based on routine blood analysis features. Med Biol Eng Comput 58, 1583–1601 (2020). https://doi.org/10.1007/s11517-020-02187-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-020-02187-9