Abstract
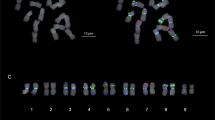
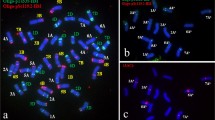
Fluorescence in situ hybridization (FISH) was applied to somatic chromosomes preparations of Oryza officinalis Wall. (CC), O. sativa L. (AA)×O. officinalis F1 hybrid (AC), backcross progenies BC1 (AAC and ACC), O. latifolia Desv. (CCDD), O. alta Swallen (CCDD) and O. punctata Kotschy (BBCC) with a labelled probe of C 0 t-1 DNA from O. officinalis. In O. officinalis, the homologous chromosomes showed similar signal bands probed by C 0 t-1 DNA and karyotype analysis was conducted based on the band patterns. Using no blocking DNA, the probe identified the chromosomes of C genome clearly, but detected few signals on chromosomes of A genome in the F1 hybrid and two backcross progenies of BC1. It is obvious that the highly and moderately repetitive DNA sequences were considerably different between C and A genomes. The chromosomes of C genome were also discriminated from the chromosomes of D-and B-genome in the tetraploid species O. latifolia, O. alta and O. punctata by C 0 t-1 DNA-FISH. Comparison of the fluorescence intensity on the chromosomes of B, C and D genomes in O. latifolia, O. alta, and O. punctata indicated that the differentiations between C and D genomes are less than that between C and B genomes. The relationship between C and D genomes in O. alta is closer than that of C and D genomes in O. latifolia. This would be one of the causes for the fact that both the genomes are of the same karyotype (CCDD) but belong to different species. The above results showed that the C 0 t-1 DNA had a high specificity of genome and species. In this paper, the origin of allotetraploid in genus Oryza is also discussed.
Similar content being viewed by others
References
Lu B R. Taxonomy of the genus Oryza (Poaceae): A historical perspective and current status. Intern Rice Resour Notes, 1999, 24: 4–8
Aggarwal R K, Brar D S, Khush G S. Two new genomes in the Oryza complex identified on the basic of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet, 1997, 254: 1–12
Ge S, Sang T, Lu B R, et al. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA, 1999, 96: 14400–14405
Lu B R, Ge S, Sang T, et al. The current taxonomy and perplexity of the genus Oryza (Poaceae). Acta Phytotaxon Sin (in Chinese), 2001, 39: 373–388
Vaughan D A. The genus Oryza L. Current status of taxonomy. IRRI Res Paper Series, 1989, 138: 1–21
Vaughan D A, Morishima H, Kadowaki K. Diversity in the Oryza genus. Curr Opin Plant Biol, 2003, 6: 139–146
Bao Y, Lu B R, Ge S. Identification of genomic constitutions of Oryza species with the B and C genomes by the PCR-RFLP method. Genet Resour Crop Ev, 2005, 52: 69–76
Jena K K, Khush G S. Introgression of genes from Oryza officinalis Well ex Watt to cultivated rice, O.sativa L. Theor Appl Genet, 1990, 80: 737–745
Kobayashi N, Ikeda R, Vaughan D A, et al. Resistance to Tungro in some wild relatives of rice. Rice Res Neusel, 1991, 16: 13
Multani D S, Khush G S, Reyes delos B G, et al. Alien genes introgression and development of monosomic alien addition lines from Oryza latifolia Desv. to rice, Oryza sativa L. Theor Appl Genet, 2003, 107: 395–405
Liu L, Lafitte R, Guan D. Wild Oryza species as potential sources of drought-adaptive traits. Euphytica, 2004, 138: 149–161
Fukui K, Shishido R, Kinoshita T. Identification of the rice D-genome chromosomes by genomic in situ hybridisation. Theor Appl Genet, 1997, 95: 1239–1245
Shishido R, Apisitiwanich S, Ohmido N, et al. Detection of specific chromosome reduction in rice somatic hybrids with the A, B, and C genomes by multi-color genomic in situ hybridization. Theor Appl Genet, 1998, 97: 1013–1018
Li C B, Zhang D M, Ge S, et al. Identification of genome constitution of Oryza malampuzhaensis, O. minuta, and O. punctataby multicolor genomic in situ hybridization. Theor Appl Genet, 2001, 103: 204–211
Li C B, Zhang D M, Ge S, et al. Differentiation and inter-genomic relationships among C, E and D genomes in the Oryza officinalis complex (Poaceae) as revealed by multicolor genomic in situ hybridization. Theor Appl Genet, 2001, 103: 197–203
Rokka V M, Clark M S, Knudson D L, et al. Cytological and molecular characterization of repetitive DNA sequences of Solanum brevidens and Solanum tuberosum. Genome, 1998, 41: 487–494
Ohmido N, Kijima K, Akiyama Y, et al. Quantification of total genomic DNA and selected repetitive sequences reveals concurrent changes in different DNA families in indica and japonica rice. Mol Gen Genet, 2000, 263: 388–394
Flavell R B, Bennett M D, Smith J B, et al. Genome size and proportion of repeated sequence DNA in plant. Biochem Genet, 1974, 12: 257–269
Bennetzen J L, Jianxin M A, Devos K M. Mechanisms of recent genome size variation in flowering plants. Ann Bot-London, 2005, 95: 127–132
McCouch S R, Tanskley S D. The world rice economy: Challenges ahead. In: Khush G S, Toenniessen G H, eds. Rice Biotechnology: Biotechnology in Agriculture Series, No. 6. Wallingford: Commonwealth Agricultural Bureaux International Press, 1991
Galasso I, Schmidt T, Pignone D, et al. The molecular cytogenetics of Vigna unguiculata (L) Walp: The physical organization and characterization of 18s-58s-25s rRNA genes, 5s rRNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theor Appl Genet, 1995, 91: 928–935
Wang Z X, Kurata N, Saji S, et al. A chromosome 5-specific repetitive DNA sequence in rice (Oryza sativa L). Theor Appl Genet, 1995, 90: 907–913
Matyasek R, Gazdova B, Fajkus J, et al. NTRS, a new family of highly repetitive DNAs specific for the T1 chromosome of tobacco. Chromosoma, 1997, 106: 369–379
Hall A E, Keith K C, Hall S E, et al. The rapidly evolving field of plant centromeres. Curr Opin Plant Biol, 2004, 7: 108–114
Linares C, Ferrer E, Fominaya A. Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L. Proc Natl Acad Sci USA, 1998, 95: 12450–12455
Cheng Z K, Stupar R M, Gu M H, et al. A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma, 2001, 110: 24–31
Cheng Z K, Buell C R, Wing R A, et al. Toward a cytological characterization of the rice genome. Genome Res, 2001, 11: 2133–2141
Cheng Z K, Dong F G, Langdon T, et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell, 2002, 14: 1691–1704
Yan H H, Liu G Q, Cheng Z K, et al. A genome-specific repetitive DNA sequence from Oryza eichingeri: Characterization, localization, and introgression to O. sativa. Theor Appl Genet, 2002, 104: 177–183
Mishima M, Ohmido N, Fukui K, et al. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma, 2002, 110: 550–558
** W W, Melo J R, Nagaki K, et al. Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell, 2004, 16: 571–581
** W W, Lamb J C, Vega J M, et al. Molecular and functional dissection of the maize B chromosome centromere. Plant Cell, 2005, 17: 1412–1423
Zwick M S, Hanson R E, Mcknight T D, et al. A rapid procedure for the isolation of C 0t-1 DNA from plants. Genome, 1997, 40: 138–142
Ren N, Song Y C, Bi X Z, et al. The physical location of genes cdc2 and prh1 in Maize (Zea mays L.). Hereditas, 1997, 126: 211–217
Doyle J J, Doyle J L. Isolation of plant DNA from fresh tissue. Focus, 1990, 12: 13–15
Jiang J M, Gill B S, Wang G L, et al. Metaphase and interphase fluorescence in situ hybridization map** of the rice genome with bacterial artificial chromosome. Proc Natl Acad Sci USA, 1995, 92: 4487–4491
Uozu S, Ikehashin H, Ohmido N, et al. Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol Biol, 1997, 35: 791–799
Bennett M D, Leitch I J. Plant genome size research: A field in focus. Ann Bot-London, 2005, 95: 1–6
Cheng Z K, Yan H H, Yu H X, et al. Development and applications of a complete set of rice telotrisomics. Genetics, 2001, 157: 161–168
Feng Q, Zhang Y, Hao P, et al. Sequence and analysis of rice chromosome 4. Nature, 2002, 420: 316–320
Ren F G, Lu B R, Li S Q, et al. A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theor Appl Genet, 2003, 108: 113–120
Zhang Y, Huang Y C, Zhang L, et al. Structural features of the rice chromosome 4 centromere. Nucleic Acids Res, 2004, 32: 2023–2030
Qin R, Wei W H, ** W W, et al. Physical location of rice Gm-6, Pi-5(t) genes in O. officinalis with BAC-FISH. Chin Sci Bull, 2001, 46: 2427–2430
Yan H H, Cheng Z K, Liu G Q, et al. Identification of Oryza×Oryza officinalis F1 and backcross progenies using genomic in situ hybridization. Acta Genet Sin (in Chinese), 1999, 26: 157–162
Feuillet C, Keller B. High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA, 1999, 96: 8265–8270
Choi H K, Mun J H, Kim D J, et al. Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA, 2004, 101: 15289–15294
Schwarzacher T, Leitch A R, Bennett M D, et al. In situ hybridization of parental genomes in a wide hybrid. Ann Bot, 1989, 64: 315–324
Ku H M, Vision T, Liu J P, et al. Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA, 2000, 97: 9121–9126
Multani D S, Khush G S, Reyes B G, et al. Alien genes introgression and development of monosomic alien addition lines from Oryza latifolia Desv. to rice, Oryza sativa L. Theor Appl Genet, 2003, 107: 395–405
Yin P, Hartemink A J. Theoretical and practical advances in genome halving. Bioinformatics, 2005, 21: 869–879
Yogeeswaran K, Frary A, York T L, et al. Comparative genome analyses of Arabidopsis spp.: Inferring chromosomal rearrangement events in the evolutionary history of A.thaliana. Genome Res, 2005, 15: 505–515
Jena K K, Kochert G. Restriction fragment length polymorphism analysis of CCDD genome species of the genus Oryza L. Plant Mol Bio, 1991, 16: 831–839
Wang Z Y, Second G, Tanksley S D. Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet, 1992, 83: 565–581
Lim K Y, Leitch I J, Leitch A R. Genomic characterization and the detection of raspberry chromatin in polyploid Rubus. Theor Appl Genet, 1998, 97: 1027–1033
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
About this article
Cite this article
Lan, W., Qin, R., Li, G. et al. Comparative analysis of A, B, C and D genomes in the genus Oryza with C 0 t-1 DNA of C genome. CHINESE SCI BULL 51, 1710–1720 (2006). https://doi.org/10.1007/s11434-006-2049-5
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11434-006-2049-5