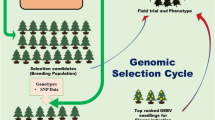
Abstract
For the most part, molecular markers and detection of quantitative trait loci have been developed for forest tree species in view to performing marker-assisted selection (MAS). However, MAS has not been applied to forest trees until now. In parallel, some success stories of MAS in crop breeding have been reported. Recently, genoty** techniques have undergone a tremendous increase in throughput, moving the trend from MAS to genomic selection. We analyzed 250 papers reporting the use of MAS in plant breeding and found that the most popular schemes used were gene pyramiding and marker-assisted backcross manipulating a single or very few genomic regions which have a major impact on crop value. We reviewed theoretical and simulation studies to identify the parametric space in which MAS is expected to bring about significant advantages over phenotypic selection. Then, we tried to explain why MAS has not been applied to forest trees and discuss the opportunities offered by recent advances in these species.




Similar content being viewed by others
References
Ahmadi N, Albar L, Pressoir G et al (2001) Genetic basis and map** of the resistance to Rice yellow mottle virus. III. Analysis of QTL efficiency in introgressed progenies confirmed the hypothesis of complementary epistasis between two resistance QTLs. Theor Appl Genet 103:1084–1092
Alves de Moraes RM, Bastos Soares TC, Colombo LR et al (2006) Assisted selection by specific DNA markers for genetic elimination of the kunitz trypsin inhibitor and lectin in soybean seeds. Euphytica 149:221–226. doi:10.1007/s10681-005-9069-0
Babu R, Nair SK, Kumar A et al (2005) Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM). Theor Appl Genet 111:888–897. doi:10.1007/s00122-005-0011-6
Basavaraj SH, Singh VK, Atul S et al (2010) Marker-assisted improvement of bacterial blight resistance in parental lines of Pusa RH10, a superfine grain aromatic rice hybrid. Mol Breed 26:293–305
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664. doi:10.2135/cropsci2008.03.0131
Bernardo RC, Charcosset A (2006) Usefulness of gene information in marker-assisted recurrent selection: a simulation appraisal. Crop Sci 46:614–621. doi:10.2135/cropsci2005.05-0088
Bhatia D, Sharma R, Vikal Y et al (2011) Marker-assisted development of bacterial blight resistant, dwarf, and high yielding versions of two traditional basmati rice cultivars. Crop Sci 51:759–770. doi:10.2135/cropsci2010.06.0358
Birol I, Raymond A, Jackman SD et al (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29:1492–1497. doi:10.1093/bioinformatics/btt178
Blanc G, Charcosset A, Veyrieras JB et al (2008) Marker-assisted selection efficiency in multiple connected populations: a simulation study based on the results of a QTL detection experiment in maize. Euphytica 161:71–84
Boichard D, Guillaume F, Baur A et al (2012) Genomic selection in French dairy cattle. Anim Prod Sci 52:115–120
Burdon RD, Wilcox PL (2007) Population management: potential impacts of advances in genomics. New For 34:187–206. doi:10.1007/s11056-007-9047-6
Causse M, Chaib J, Lecomte L et al (2007) Both additivity and epistasis control the genetic variation for fruit quality traits in tomato. Theor Appl Genet 115:429–442. doi:10.1007/s00122-007-0578-1
Chain P, Grafham D, Fulton R et al (2009) Genome project standards in a new era of sequencing. Science 326:236
Chakraborty R, Moreau L, Dekkers JC (2002) A method to optimize selection on multiple identified quantitative trait loci. Genet Sel Evol 34:145–170. doi:10.1186/1297-9686-34-2-145
Chancerel E, Lepoittevin C, Provost GL et al (2011) Development and implementation of a highly-multiplexed SNP array for genetic map** in maritime pine and comparative map** with loblolly pine. BMC Genomics 12:368. doi:10.1186/1471-2164-12-368
Chen S, Lin XH, Xu CG, Zhang Q (2000) Improvement of bacterial blight resistance of “Minghui 63”, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Sci 40:239–244
Chen S, Xu CG, Lin XH, Zhang Q (2001) Improving bacterial blight resistance of “6078”, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Plant Breed 120:133–137. doi:10.1046/j.1439-0523.2001.00559.x
Chen CL, Chuang SJ, Chen JJ, Sung JM (2009) Using RAPD markers to predict polyphenol content in aerial parts of Echinacea purpurea plants. J Sci Food Agric 89:2137–2143. doi:10.1002/jsfa.3704
Chen C, Mitchell SE, Elshire RJ et al (2013) Mining conifers’ mega-genome using rapid and efficient multiplexed high-throughput genoty**-by-sequencing (GBS) SNP discovery platform. Tree Genet Genomes 9:1537–1544. doi:10.1007/s11295-013-0657-1
Cornelius J (1994) Heritabilities and additive genetic coefficients of variation in forest trees. Can J For Res 24:372–379. doi:10.1139/x94-050
da Silva FF, Pereira MG, Campos WF et al (2007) DNA marker-assisted sex conversion in elite papaya genotype (Carica papaya L.). Crop Breed Appl Biotechnol 7:52–58
Darvasi A, Soller M (1997) A simple method to calculate resolving power and confidence interval of QTL map location. Behav Genet 27:125–132. doi:10.1023/A:1025685324830
Dekkers JCM (2007) Prediction of response to marker-assisted and genomic selection using selection index theory. J Anim Breed Genet 124:331–341
Dekkers JC, Chakraborty R, Moreau L (2002) Optimal selection on two quantitative trait loci with linkage. Genet Sel Evol 34:171–192. doi:10.1186/1297-9686-34-2-171
Devey ME, Groom KA, Nolan MF et al (2004) Detection and verification of quantitative trait loci for resistance to Dothistroma needle blight in Pinus radiata. Theor Appl Genet 108:1056–1063. doi:10.1007/s00122-003-1471-1
Dolstra O, Denneboom C, Vos ALF de, Loo EN van (2007) Marker-assisted selection for improving quantitative traits of forage crops. In: Guimarães EP, Ruane J, Scherf BD, Sonnino A, Dargie JD (eds) Marker-assisted selection: current status and future perspectives in crops, livestock, forestry and fish ed. Food and Agriculture Organization of the United Nations (FAO), Rome Italy, pp. 59–65
Eckert AJ, van Heervaarden J, Wegrzyn JL et al (2010) Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae). Genetics 185:969–982. doi:10.1534/genetics.110.115543
Edwards MD, Page NJ (1994) Evaluation of marker-assisted selection through computer simulation. Theor Appl Genet 88:376–382
El-Kassaby YA, Lstiburek M (2009) Breeding without breeding. Genet Res 91:111–120. doi:10.1017/S001667230900007X
El-Kassaby YA, Cappa EP, Liewlaksaneeyanawin C et al (2011) Breeding without breeding: is a complete pedigree necessary for efficient breeding? PLoS ONE 6:e25737. doi:10.1371/journal.pone.0025737
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genoty**-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. doi:10.1371/journal.pone.0019379
Emebiri L, Michael P, Moody DB et al (2009a) Pyramiding QTLs to improve malting quality in barley: gains in phenotype and genetic diversity. Mol Breed 23:219–228. doi:10.1007/s11032-008-9227-x
Emebiri LC, Michael P, Moody DB (2009b) Enhanced tolerance to boron toxicity in two-rowed barley by marker-assisted introgression of favourable alleles derived from Sahara 3771. Plant Soil 314:77–85. doi:10.1007/s11104-008-9707-0
Fernando RL, Grossman M (1989) Marker assisted selection using best linear unbiased prediction. Genet Sel Evol 21:467–477
Feuillet C, Leach JE, Rogers J et al (2011) Crop genome sequencing: lessons and rationales. Trends Plant Sci 16:77–88. doi:10.1016/j.tplants.2010.10.005
Flachowsky H, Le Roux P-M, Peil A et al (2011) Application of a high-speed breeding technology to apple (Malus × domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytol 192:364–377. doi:10.1111/j.1469-8137.2011.03813.x
Flint-Garcia SA, Darrah LL, McMullen MD, Hibbard BE (2003a) Phenotypic versus marker-assisted selection for stalk strength and second-generation European corn borer resistance in maize. Theor Appl Genet 107:1331–1336. doi:10.1007/s00122-003-1387-9
Flint-Garcia SA, Thornsberry JM, Buckler SE IV (2003b) Structure of linkage disequilibrium in plants. Annual Review of Plant Biology 54:357–374. doi:10.1146/annurev.arplant.54.031902.134907
Gill GP, Harvey CF, Gardner RC, Fraser LG (1998) Development of sex-linked PCR markers for gender identification in Actinidia. Theor Appl Genet 97:439–445. doi:10.1007/s001220050914
Goddard ME (2001) The validity of genetic models underlying quantitative traits. Livest Prod Sci 72:117–127
Grattapaglia D (2007) Marker-assisted selection in Eucalyptus. In: Guimaraes EP, Ruane J, Scherf BD (eds) Marker-assisted selection: current status and future perspectives in crops, livestock, forestry and fish ed. Food and Agriculture Organization of the United Nations (FAO), Rome, pp 251–281
Grattapaglia D, Kirst M (2008) Eucalyptus applied genomics: from gene sequences to breeding tools. New Phytol 179:911–929. doi:10.1111/j.1469-8137.2008.02503.x
Grattapaglia D, Resende MDV (2011) Genomic selection in forest tree breeding. Tree Genet Genom 7:241–255
Guillaume F, Fritz S, Boichard D, Druet T (2008) Estimation by simulation of the efficiency of the French marker-assisted selection program in dairy cattle. Genet Sel Evol 40:91–102. doi:10.1051/gse:2007036
Gupta P, Rustgi S, Kulwal P (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol 57:461–485. doi:10.1007/s11103-005-0257-z
Hamberger B, Hall D, Yuen M et al (2009) Targeted isolation, sequence assembly and characterization of two white spruce (Picea glauca) BAC clones for terpenoid synthase and cytochrome P450 genes involved in conifer defence reveal insights into a conifer genome. BMC Plant Biol 9:106. doi:10.1186/1471-2229-9-106
Han F, Romagosa I, Ullrich SE et al (1997) Molecular marker-assisted selection for malting quality traits in barley. Mol Breed 3:427–437
Harushima Y, Yano M, Shomura P et al (1998) A high-density rice genetic linkage map with 2275 markers using a single F-2 population. Genetics 148:479–494
Hayes B, Baranski M, Goddard ME, Robinson N (2007) Optimisation of marker assisted selection for abalone breeding programs. Aquaculture 265:61–69
Hill W, Weir B (2011) Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet Res 93:47–64
Hirakawa H, Nakamura Y, Kaneko T et al (2011) Survey of the genetic information carried in the genome of Eucalyptus camaldulensis. Plant Biotechnol 28:471–480
Hospital F (2009) Challenges for effective marker-assisted selection in plants. Genetica 136:303–310. doi:10.1007/s10709-008-9307-1
Hospital F, Charcosset A (1997) Marker-assisted introgression of quantitative trait loci. Genetics 147:1469–1485
Huang N, Angeles ER, Domingo J et al (1997) Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor Appl Genet 95:313–320. doi:10.1007/s001220050565
Hyten DL, Choi IY, Song QJ et al (2010) A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus map**. Crop Sci 50:960–968. doi:10.2135/cropsci2009.06.0360
Igartua E, Edney M, Rossnagel BG et al (2000) Marker-based selection of QTL affecting grain and malt quality in two-row barley. Crop Sci 40:1426–1433. doi:10.2135/cropsci2000.4051426x
Iwata H, Hayashi T, Tsumura Y (2011) Prospects for genomic selection in conifer breeding: a simulation study of Cryptomeria japonica. Tree Genet Genom 7:747–758. doi:10.1007/s11295-011-0371-9
Jena KK, Mackill DJ (2008) Molecular markers and their use in marker-assisted selection in rice. Crop Sci 48:1266–1276. doi:10.2135/cropsci2008.02.0082
Jermstad KD, Bassoni DL, Jech KS et al (2001) Map** of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. I. Timing of vegetative bud flush. Theor Appl Genet 102:1142–1151. doi:10.1007/s001220000505
Johnson GR, Wheeler NC, Strauss SH (2000) Financial feasibility of marker-aided selection in Douglas-fir. Can J For Res 30:1942–1952. doi:10.1139/x00-122
Jonas E, de Koning D-J (2013) Does genomic selection have a future in plant breeding? Trends Biotechnol 31:497–504. doi:10.1016/j.tibtech.2013.06.003
Kearsey MJ, Farquhar AGL (1998) QTL analysis in plants; where are we now? Heredity 80:137–142. doi:10.1046/j.1365-2540.1998.00500.x
Kuchel H, Ye G, Fox R, Jefferies S (2005) Genetic and economic analysis of a targeted marker-assisted wheat breeding strategy. Mol Breed 16:67–78. doi:10.1007/s11032-005-4785-7
Kumar S, Garrick DJ (2001) Genetic response to within-family selection using molecular markers in some radiata pine breeding schemes. Can J For Res 31:779–785
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124:743–756
Levi A, Paterson A, Barak V et al (2009) Field evaluation of cotton near-isogenic lines introgressed with QTLs for productivity and drought related traits. Mol Breed 23:179–195. doi:10.1007/s11032-008-9224-0
Lorenz AJ, Chao S, Asoro FG et al (2011) Genomic selection in plant breeding: knowledge and prospects. In: Sparks DL (ed) Advances in agronomy, Vol 110th edn. Elsevier, San Diego, pp 77–123
Mahmood T, Rahman MH, Stringam GR et al (2005) Molecular markers for yield components in Brassica juncea—do these assist in breeding for high seed yield? Euphytica 144:157–167. doi:10.1007/s10681-005-5339-0
Massah N, Wang J, Russell JH et al (2010) Genealogical relationship among members of selection and production populations of yellow cedar (Callitropsis nootkatensis [D. Don] Oerst.) in the absence of parental information. J Hered 101:154–163. doi:10.1093/jhered/esp102
Mayor PJ, Bernardo R (2009) Doubled haploids in commercial maize breeding: one-stage and two-stage phenotypic selection versus marker-assisted recurrent selection. Maydica 54:439–448
Meuwissen T, Hayes B, Goddard M (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Moreau L, Charcosset A, Hospital F, Gallais A (1998) Marker-assisted selection efficiency in populations of finite size. Genetics 148:1353–1365
Moreau L, Charcosset A, Gallais A (2004) Experimental evaluation of several cycles of marker-assisted selection in maize. Euphytica 137:111–118. doi:10.1023/b:euph.0000040508.01402.21
Morris M, Dreher K, Ribaut JM, Khairallah M (2003) Money matters (II): costs of maize inbred line conversion schemes at CIMMYT using conventional and marker-assisted selection. Mol Breed 11:235–247. doi:10.1023/a:1022872604743
Myint Y, Khin Than N, Vanavichit A et al (2009) Marker assisted backcross breeding to improve cooking quality traits in Myanmar rice cultivar Manawthukha. Field Crops Res 113:178–186. doi:10.1016/j.fcr.2009.05.006
Neale DB, Savolainen O (2004) Association genetics of complex traits in conifers. Trends Plant Sci 9:325–330. doi:10.1016/j.tplants.2004.05.006
Neale DB, Wegrzyn JL, Stevens KA et al (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15:R59. doi:10.1186/gb-2014-15-3-r59
Nielsen HM, Sonesson AK, Meuwissen THE (2011) Optimum contribution selection using traditional best linear unbiased prediction and genomic breeding values in aquaculture breeding schemes. J ANIM SCI 89:630–638. doi:10.2527/jas.2009-2731
Nystedt B, Street NR, Wetterbom A et al (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. doi:10.1038/nature12211
Ollivier L (1998) The accuracy of marker-assisted selection for quantitative traits within populations in linkage equilibrium. Genetics 148:1367–1372
Parasnis AS, Ramakrishna W, Chowdari KV et al (1999) Microsatellite (GATA)n reveals sex-specific differences in papaya. Theor Appl Genet 99:1047–1052. doi:10.1007/s001220051413
Parchman TL, Gompert Z, Mudge J et al (2012) Genome-wide association genetics of an adaptive trait in lodgepole pine. Mol Ecol 21:2991–3005. doi:10.1111/j.1365-294X.2012.05513.x
Perumalsamy S, Bharani M, Sudha M et al (2010) Functional marker-assisted selection for bacterial leaf blight resistance genes in rice (Oryza sativa L.). Plant Breed 129:400–406
Plomion C, Durel CE, Verhaegen D (1996) Marker-assisted selection in forest tree breeding programs as illustrated by two examples: maritime pine and eucalyptus. Ann For Sci 53:819–848
Plomion C, LeProvost G, Pot D et al (2001) Pollen contamination in a maritime pine polycross seed orchard and certification of improved seeds using chloroplast microsatellites. Can J For Res 31:1816–1825. doi:10.1139/cjfr-31-10-1816
Prasanna BM, Pixley K, Warburton ML, **e C-X (2010) Molecular marker-assisted breeding options for maize improvement in Asia. Mol Breed 26:339–356. doi:10.1007/s11032-009-9387-3
Prat D, Faivre-Rampant P, Prado E (2006) Genome analysis and the management of forest genetic resources. Institut National de la Recherche Agronomique (INRA), Paris
Pryce JE, Hayes BJ, Goddard ME (2012) Novel strategies to minimize progeny inbreeding while maximizing genetic gain using genomic information. J Dairy Sci 95:377–388. doi:10.3168/jds.2011-4254
Resende MFR Jr, Muñoz P, Acosta JJ et al (2012a) Accelerating the domestication of trees using genomic selection: accuracy of prediction models across ages and environments. New Phytol 193:617–624. doi:10.1111/j.1469-8137.2011.03895.x
Resende MFR Jr, Muñoz P, Resende MDV et al (2012b) Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 190:1503–1510. doi:10.1534/genetics.111.137026
Resende MDV, Resende MFR Jr, Sansaloni CP et al (2012c) Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol 194:116–128. doi:10.1111/j.1469-8137.2011.04038.x
Ribeiro MM, Sanchez L, Ribeiro C et al (2011) A case study of Eucalyptus globulus fingerprinting for breeding. Ann For Sci 68:701–714. doi:10.1007/s13595-011-0087-x
Rosvall O (2011) Review of the Swedish breeding programme. Skogforsk, Uppsala, Sweden
Sánchez L, Caballero A, Santiago E (2006) Palliating the impact of fixation of a major gene on the genetic variation of artificially selected polygenes. Genet Res 88:105–118. doi:10.1017/S0016672306008421
Sebastian SA, Streit LG, Stephens PA et al (2010) Context-specific marker-assisted selection for improved grain yield in elite soybean populations. Crop Sci 50:1196–1206. doi:10.2135/cropsci2009.02.0078
Sewell M, Bassoni D, Megraw R et al (2000) Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). I. Physical wood properties. Theor Appl Genet 101:1273–1281
Sharma K, Agrawal V, Gupta S et al (2008) ISSR marker-assisted selection of male and female plants in a promising dioecious crop: jojoba (Simmondsia chinensis). Plant Biotechnol Rep 2:239–243. doi:10.1007/s11816-008-0070-7
Sillanpää MJ (2011) On statistical methods for estimating heritability in wild populations. Mol Ecol 20:1324–1332. doi:10.1111/j.1365-294X.2011.05021.x
Silva KM, Bastiaansen JWM, Knol EF et al (2011) Meta-analysis of results from quantitative trait loci map** studies on pig chromosome 4. Anim Genet 42:280–292. doi:10.1111/j.1365-2052.2010.02145.x
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Sorkheh K, Malysheva-Otto LV, Wirthensohn MG et al (2008) Linkage disequilibrium, genetic association map** and gene localization in crop plants. Genet Mol Biol 31:805–814. doi:10.1590/S1415-47572008005000005
Spelman R, Bovenhuis H (1998) Genetic response from marker assisted selection in an outbred population for differing marker bracket sizes and with two identified quantitative trait loci. Genetics 148:1389–1396
Steele KA, Price AH, Shashidhar HE, Witcombe JR (2006) Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor Appl Genet 112:208–221
Stendal C, Casler MD, Jung G (2006) Marker-assisted selection for neutral detergent fiber in smooth bromegrass. Crop Sci 46:303–311
Stock K, Reents R (2013) Genomic selection: status in different species and challenges for breeding. Reprod Domest Anim 48:2–10. doi:10.1111/rda.12201
Szücs PB, Bhat VC, Chao PR et al (2009) An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome 2:134
Tanhuanpää P, Vilkki J (1999) Marker-assisted selection for oleic acid content in spring turnip rape. Plant Breed 118:568–570
Tanksley SD, Young ND, Paterson AH, Bonierbale MW (1989) RFLP map** in plant breeding: new tools for an old science. Nat Biotechnol 7:257–264. doi:10.1038/nbt0389-257
Truntzler M, Barrière Y, Sawkins M et al (2010) Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes. Theor Appl Genet 121:1465–1482. doi:10.1007/s00122-010-1402-x
Twardowska M, Masojc P, Milczarski P (2005) Pyramiding genes affecting sprouting resistance in rye by means of marker assisted selection. In:(ed) Proceedings of the 10th International Symposium on Pre-Harvest Sprouting in Cereals, Norfolk, UK, 7–11 June 2004. ed. Norfolk, UK, pp 257–260
Van de Wen WTG, McNicol RJ (1995) The use of RAPD markers for the identification of Sitka spruce (Picea sitchensis) clones. Heredity 75:126–132
van der Knaap E, Lippman ZB, Tanksley SD (2002) Extremely elongated tomato fruit controlled by four quantitative trait loci with epistatic interactions. Theor Appl Genet 104:241–247. doi:10.1007/s00122-001-0776-1
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423. doi:10.3168/jds.2007-0980
Varshney R, Marcel T, Ramsay L et al (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet 114:1091–1103. doi:10.1007/s00122-007-0503-7
Veyrieras J-B, Goffinet B, Charcosset A (2007) MetaQTL: a package of new computational methods for the meta-analysis of QTL map** experiments. BMC Bioinformatics 8:49. doi:10.1186/1471-2105-8-49
Vida G, Gal M, Uhrin A et al (2009) Molecular markers for the identification of resistance genes and marker-assisted selection in breeding wheat for leaf rust resistance. Euphytica 170:67–76. doi:10.1007/s10681-009-9945-0
Villanueva B, Pong-Wong R, Woolliams JA (2002) Marker assisted selection with optimised contributions of the candidates to selection. Genet Sel Evol 34:679–703. doi:10.1051/gse:2002031
Visscher PM, Hill WG, Wray NR (2008) Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet 9:255–266. doi:10.1038/nrg2322
Wilcox PL, Carson SD, Richardson TE et al (2001) Benefit-cost analysis of DNA marker-based selection in progenies of Pinus radiata seed orchard parents. Can J For Res 31:2213–2224. doi:10.1139/cjfr-31-12-2213
Wong CK, Bernardo R (2008) Genomewide selection in oil palm: increasing selection gain per unit time and cost with small populations. Theor Appl Genet 116:815–824. doi:10.1007/s00122-008-0715-5
**e C, Xu S (1998) Efficiency of multistage marker-assisted selection in the improvement of multiple quantitative traits. Heredity 80:489–498. doi:10.1038/sj.hdy.6883080
Xu SZ (2003) Theoretical basis of the Beavis effect. Genetics 165:2259–2268
Xu YB, Crouch JH (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48:391–407. doi:10.2135/cropsci2007.04.0191
Zimin A, Stevens KA, Crepeau MW et al (2014) Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics 196:875–890. doi:10.1534/genetics.113.159715
Acknowledgments
This work was funded by the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 211868 (Project Noveltree).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. G. Abbott
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Distribution of marker density in 48 papers, concerning 11 species, reporting genetic maps in forest trees. (PNG 52 kb)
Supplementary document 1
A text explaining how we selected articles for the meta-analysis and a table showing the queries used and the number of articles obtained. (PDF 10 kb)
Supplementary document 2
List of references of the articles used in the meta-analysis, in RIS format. (PDF 626 kb)
Supplementary document 3
List of references of the articles cited in Tables 2 and 3, in RIS format. (PDF 117 kb)
Rights and permissions
About this article
Cite this article
Muranty, H., Jorge, V., Bastien, C. et al. Potential for marker-assisted selection for forest tree breeding: lessons from 20 years of MAS in crops. Tree Genetics & Genomes 10, 1491–1510 (2014). https://doi.org/10.1007/s11295-014-0790-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0790-5