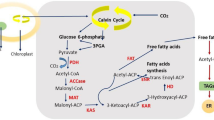
Abstract
Biomass production is currently explored in microalgae, macroalgae and land plants. Microalgal biofuel development has been performed mostly in green algae. In the Japanese tradition, macrophytic red algae such as Pyropia yezoensis and Gelidium crinale have been utilized as food and industrial materials. Researches on the utilization of unicellular red microalgae such as Cyanidioschyzon merolae and Porphyridium purpureum started only quite recently. Red algae have relatively large plastid genomes harboring more than 200 protein-coding genes that support the biosynthetic capacity of the plastid. Engineering the plastid genome is a unique potential of red microalgae. In addition, large-scale growth facilities of P. purpureum have been developed for industrial production of biofuels. C. merolae has been studied as a model alga for cell and molecular biological analyses with its completely determined genomes and transformation techniques. Its acidic and warm habitat makes it easy to grow this alga axenically in large scales. Its potential as a biofuel producer is recently documented under nitrogen-limited conditions. Metabolic pathways of the accumulation of starch and triacylglycerol and the enzymes involved therein are being elucidated. Engineering these regulatory mechanisms will open a possibility of exploiting the full capability of production of biofuel and high added-value oil. In the present review, we will describe the characteristics and potential of these algae as biotechnological seeds.



Similar content being viewed by others
References
Araki S, Sakurai T, Omata T, Kawaguchi A, Murata N (1986) Lipid and fatty acid composition in the red alga Porphyra yezoensis. Jap. J Phycol 34:94–100
Araki S, Sakurai T, Kawaguchi A, Murata N (1987) Positional distribution of fatty acids in glycerolipids of the marine red alga, Porphyra yezoensis. Plant Cell Physiol 28:761–766
Araki S, Sakurai T, Oohusa T, Kayama M, Sato N (1989) Characterization of sulfoquinovosyl diacylglycerol from marine red algae. Plant Cell Physiol 30:775–781
Araki S, Sakurai T, Oohusa T, Kayama M, Nisizawa K (1990) Content of arachidonic and eicosapentaenoic acids in polar lipids from Gracilaria (Gracilariales, Rhodophyta). Hydrobiologia 204/205:513–519
Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C (2011) The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J Exp Bot 62:1775–1801
Bates PD (2016) Understanding the control of the acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim Biophys Acta 1861:1214–1225
Bhattacharya D, Price DC, Chan CX, Qiu H, Rose N, Ball S, Weber AP, Arias MC, Henrissat B, Coutinho PM, Krishnan A, Zäuner S, Morath S, Hilliou F, Egizi A, Perrineau MM, Yoon HS (2013) Genome of the red alga Porphyridium purpureum. Nat Commun 4:1941
Cavalier-Smith T (2003) Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote–eukaryote chimaeras (meta-algae). Phil Trans R Soc Lond B 358:109–134
Ciniglia C, Yoon HS, Pollio A, Pinto G, Bhattacharya D (2004) Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol Ecol 13:1827–1837
Collén J, Porcel B, Carré W, Ball SG, Chaparro C, Tonon T, Barbeyron T, Michel G, Noel B, Valentin K, Elias M, Artiguenave F, Arun A, Aury JM, Barbosa-Neto JF, Bothwell JH, Bouget FY, Brillet L, Cabello-Hurtado F, Capella-Gutiérrez S, Charrier B, Cladière L, Cock JM, Coelho SM, Colleoni C, Czjzek M, Da Silva C, Delage L, Denoeud F, Deschamps P, Dittami SM, Gabaldón T, Gachon CM, Groisillier A, Hervé C, Jabbari K, Katinka M, Kloareg B, Kowalczyk N, Labadie K, Leblanc C, Lopez PJ, McLachlan DH, Meslet-Cladiere L, Moustafa A, Nehr Z, Nyvall Collén P, Panaud O, Partensky F, Poulain J, Rensing SA, Rousvoal S, Samson G, Symeonidi A, Weissenbach J, Zambounis A, Wincker P, Boyen C (2013) Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc Natl Acad Sci USA 110:5247–5252
De Bhowmick G, Koduru L, Sen R (2015) Metabolic pathway engineering towards enhancing microalgal lipid biosynthesis for biofuel application—A review. Renew Sustain Energy Rev 50:1239–1253
Dembitsky VM (1996) Betaine ether-linked glycerolipids: chemistry and biology. Prog Lipid Res 35:1–51
Fleck-Schneider P, Lehr F, Posten C (2007) Modelling of growth and product formation of Porphyridium purupureum. J Biotechnol 132:134–141
Fujiwara T, Ohnuma M, Yoshida M, Kuroiwa T (2013) Gene targeting in the red alga Cyanidioschyzon merolae: Single- and multi-copy insertion using authentic and chimeric selection markers. PLOS ONE 8:e73608
Gao J, Ajjawi I, Manoli A, Sawin A, Xu C, Froehlich JE, Last RL, Benning C (2009) FATTY ACID DESATURASE 4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J 60:832–839
Geresh S, Arad S, Levy-Ontman O, Zhang W, Tekoah Y, Glaser R (2009) Isolation and characterization of poly- and oligosaccharides from the red microalga Porphyridium sp. Carbohydrate Res 344:343–349
Guschina IA, Harwood JL (2006) Lipids and lipid metabolism in eukaryotic algae. Prog Lipid Res 45:160–186
Hirashima T, Tajima N, Sato N (2016) Draft genome sequences of four species of Chlamydomonas containing phosphatidylcholine. Genome Announc 4:e01070–16
Iwai M, Hori K, Sasaki-Sekimoto Y, Shimojima M, Ohta H (2015) Manipulation of oil synthesis in Nannochloropsis strain NIES-2145 with a phosphorus starvation-inducible promoter from Chlamydomonas reinhardtii. Front Microbiol 6:912
Karpagam R, Preeti R, Ashokkumar B, Varalakshmi P (2015) Enhancement of lipid production and fatty acid profiling in Chlamydomonas reinhardtii, CC1010 for biodiesel production. Ecotoxicol Environ Saf 121:253–257
Kayama M, Imayoshi J, Araki S, Ogawa H, Oohusa T, Ueno T, Saito M (1983) Changes in the lipids of dried laver “Nori” at different water activities. Bull Japan Soc Sci. Fisheries 49:787–793 (AbstractEnglish)
Khozin I, Adlerstein D, Bigongo C, Heimer YM, Cohen Z (1997) Elucidation of the biosynthesis of eicosapentaenoic acid in the microalga Porphyridium cruentum II. Studies with radiolabeled precursors. Plant Physiol 114:223–230
Khozin-Goldberg I, Yu HZ, Adlerstein D, Didi-Cohen S, Heimer YM, Cohen Z (2000) Triacylglycerols of the red microalga Porphyridium cruentum can contribute to the biosynthesis of eukaryotic galactolipids. Lipids 35:881–889
Lapidot M, Raveh D, Sivan A, Arad S, Shapira M (1999) Molecular analysis of the AhaS gene of Porphyridium sp. (Rhodophyta) and of a mutant resistant to sulfometuron methyl. J Phycol 35:1233–1236
Lapidot M, Raveh D, Sivan A, Arad S, Shapira M (2002) Stable chloroplast transformation of the unicellular red alga Porphyridium species. Plant Physiol 129:7–12
Li-Beisson Y, Beisson F, Riekhof W (2015) Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J 82:504–522
Matsuzaki M, Misumi O, Shin-i T, Maruyama S, Takahara M, Miyagishima S, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657
Melo T, Alves E, Azevedo V, Martins AS, Neves B, Domingues P, Calado R, Abreu MH, Domingues MR (2015) Lipidomics as a new approach for the bioprospecting of marine macroalgae—unraveling the polar lipid and fatty acid composition of Chondrus crispus. Algal Res 8:181–191
Menetrez MY (2012) An overview of algae biofuel production and potential environmental impact. Environ Sci Technol 46:7073–7085
Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J (2012) TAG, You’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol 23:352–363
Merchuk JC, Ronen M, Giris S, Arad S (1998) Light/dark cycles in the growth of the red microalga Porphyridium sp. Biotechnol Bioeng 59:705–713
Misra N, Panda PK, Parida BK, Mishra BK (2012) Phylogenomic study of lipid genes involved in microalgal biofuel production—candidate gene mining and metabolic pathway analyses. Evol Bioinformatics 8:545–564
Mori N, Moriyama T, Toyoshima M, Sato N (2016) Construction of global acyl lipid metabolic map by comparative genomics and subcellular localization analysis in the red alga Cyanidioschyzon merolae. Front Plant Sci 7:958
Moriyama T, Sakurai K, Sekine K, Sato N (2014a) Subcellular distribution of central carbohydrate metabolism pathways in the red alga Cyanidioschyzon merolae. Planta 240:585–598
Moriyama T, Tajima N, Sekine K, Sato N (2014b) Localization and phylogenetic analysis of enzymes related to organellar genome replication in the unicellular rhodophyte Cyanidioschyzon merolae. Genome Biol Evol 6:228–237
Naumann I, Darsow KH, Walter C, Lange HA, Buchholz R (2007) Identification of sulfoglycolipids from the alga Porphyridium purpureum by matrix-assisted laser desorption/ionisation quadrupole ion trap time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 21:3185–3192
Nichols BW, Appleby RS (1969) The distribution and biosynthesis of arachidonic acid in algae. Phytochemistry 8:1907–1915
Nozaki H, Takano H, Misumi O, Terasawa K, Matsuzaki M, Maruyama S, Nishida K, Yagisawa F, Yoshida Y, Fujiwara T, Takio S, Tamura K, Chung SJ, Nakamura S, Kuroiwa H, Tanaka K, Sato N, Kuroiwa T (2007) A 100%-complete sequence reveals unusually simple genomic features in the hot spring red alga Cyanidioschyzon merolae. BMC Biol 5:28
Oh SH, Han JG, Kim Y, Ha JH, Kim SS, Jeong MH, Jeong HS, Kim NY, Cho JS, Yoon WB, Lee SY, Kang DH, Lee HY (2009) Lipid production in Porphyridium cruentum grown under different culture conditions. J Biosci Bioeng 108:429–434
Ohnuma M, Yokoyama T, Inouye T, Sekine Y, Kuroiwa T, Tanaka K (2014) Optimization of polyethylene glycol (PEG)-mediated DNA introduction conditions for transient gene expression in the unicellular red alga Cyanidioschyzon merolae. J Gen Appl Microbiol 60:156–159
Ohta S, Chang T, Aozasa O, Kondo M, Miyata H (1992) Sustained production of arachidonic and eicosapentaenoic acids by the red alga Porphyridium purpureum cultured in a light/dark cycle. J Ferment Bioeng 74:398–402
Pal D, Khozin-Goldberg I, Cohen Z (2011) The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl Microbiol Biotechnol 90:1429–1441
Pettitt TR, Jones AL, Harwood JL (1989) Lipids of the marine red algae, Chondrus crispus and Polysiphonia lanosa. Phytochemistry 28:399–405
Qin S, Lin H, Jiang P (2012) Advances in genetic engineering of marine algae. Biotechnol Adv 30:1602–1613
Riekhof WR, Sears B, Benning C (2005) Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot Cell 4:242–252
Rodolfi L, Chini ZG, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR (2009) Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng 102:100–112
Sakurai K, Moriyama T, Sato N (2014) Detailed identification of fatty acid isomers sheds light on the probable precursors of triacylglycerol accumulation in photoautotrophically grown Chlamydomonas reinhardtii. Eukaryot Cell 13:256–266
Sakurai T, Aoki M, Ju X, Ueda T, Nakamura Y, Fujiwara S, Umemura T, Tsuzuki M, Minoda A (2016) Profiling of lipid and glycogen accumulations under different growth conditions in the sulfothermophilic red alga Galdieria sulphuraria. Bioresour Technol 200:861–866
Sastre RMR (2010) Kopplung physiologischer und verfahrenstechnischer Parameter beim Wachstum and bei der Produktbildung der Rotalge Porphyridium purpureum. Dissertation, Universität Karlsruhe 2009
Sato N (1992) Betaine lipids. Bot Mag 105:185–197
Sato N (2001) Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci 6:151–156
Sato N (2009) Gclust: trans-kingdom classification of proteins using automatic individual threshold setting. Bioinformatics 25:599–605
Sato N, Moriyama T (2007) Genomic and biochemical analysis of lipid biosynthesis in the unicellular rhodophyte Cyanidioschyzon merolae: lack of plastidic desaturation pathway results in mixed pathway of galactolipid synthesis. Eukaryot Cell 6:1006–1017
Sato N, Mori N, Hirashima T, Moriyama T (2016) Diverse pathways of biosynthesis of phosphatidylcholine in algae as estimated by labeling studies and genomic sequence analysis. Plant J 87:281–292
Satyanarayana KG, Mariano AB, Vargas JVC (2011) A review on microalgae, a versatile source for sustainable energy and materials. Int J Energy Res 35:291–311
Schönknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Banfield JF, Garavito RM, Carr K, Wilkerson C, Rensing SA, Gagneul D, Dickenson NE, Oesterhelt C, Lercher MJ, Weber AP (2013) Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339:1207–1210
Shimonaga T, Konishi M, Oyama Y, Fujiwara S, Satoh A, Fujita N, Colleoni C, Buléon A, Putaux JL, Ball SG, Yokoyama A, Hara Y, Nakamura Y, Tsuzuki M (2008) Variation in storage α-glucans of the Porphyridiales (Rhodophyta). Plant Cell Physiol 49:103–116
Shiran D, Khozin I, Heimer YM, Cohen Z (1996) Biosynthesis of eicosapentaenoic acid in the microalga Porphyridium cruentum. I: the use of externally supplied fatty acids. Lipids 31:1277–1282
Siaut M, Cuiné S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylidès C, Li-Beisson Y, Peltier G (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol 11:7. doi:10.1186/1472-6750-11-7
Sumiya N, Kawase Y, Hayakawa J, Matsuda M, Nakamura M, Era A, Tanaka K, Kondo A, Hasunuma T, Imamura S, Miyagishima S (2015) Expression of cyanobacterial acyl-ACP reductase elevates the triacylglycerol level in the red alga Cyanidioschyzon merolae. Plant Cell Physiol 56:1962–1980
Tajima T, Sato S, Maruyama F, Kurokawa K, Ohta H, Tabata S, Sekine K, Moriyama T, Sato N (2014) Analysis of the complete plastid genome of the unicellular red alga Porphyridium purpureum. J Plant Res 127:389–397
Takusagawa M, Nakajima Y, Saito T, Misumi O (2016) Primitive red alga Cyanidioschyzon merolae accumulates storage glucan and triacylglycerol under nitrogen depletion. J Gen Appl Microbiol 62:111–117
Toyoshima M, Sato N (2015) High-level accumulation of triacylglycerol and starch in photoautotrophically grown Chlamydomonas debaryana NIES-2212. Plant Cell Physiol 56:2447–2456
Toyoshima M, Mori N, Moriyama T, Misumi O, Sato N (2016) Analysis of triacylglycerol accumulation under nitrogen deprivation in the red alga Cyanidioschyzon merolae. Microbiology 162:803–812
Vítová M, Goecke F, Sigler K, Řezank T (2016) Lipidomic analysis of the extremophilic red alga Galdieria sulphuraria in response to changes in pH. Algal Res 13:218–226
Zienkiewicz K, Du ZY, Ma W, Vollheyde K, Benning C (2016) Stress-induced neutral lipid biosynthesis in microalgae—molecular, cellular and physiological insights. Biochim Biophys Acta 1861:1269–1281
Acknowledgements
The studies in the authors’ laboratory have been supported in part by the CREST project on algal biotechnology from Japan Science and Technology Agency.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sato, N., Moriyama, T., Mori, N. et al. Lipid metabolism and potentials of biofuel and high added-value oil production in red algae. World J Microbiol Biotechnol 33, 74 (2017). https://doi.org/10.1007/s11274-017-2236-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-017-2236-3