Abstract
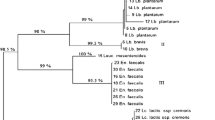
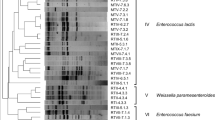
The present study aimed to evaluate the dominant microbial community naturally present in the Planalto de Bolona cheese, produced in the Cape Verde Islands. Samples of milk, curd and cheese from two different producers were examined through culture-dependent and independent-methods. Traditional plating and genetic identification of lactic acid bacteria (LAB) and yeast isolates were carried out. Moreover, DNA and RNA extracted directly from samples were subjected to Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis (PCR-DGGE). Concerning the LAB population, a total of 278 isolates were identified: Lactococcus lactis subsp. lactis and Enterococcus faecium represented the most isolated species. Regarding yeasts, the analysis of isolates throughout the manufacturing period showed a consistent presence of the genus Candida. Divergences in species detection between culture-dependent and culture-independent methods were observed, as well as between DNA and RNA analysis. PCR-DGGE underlined high heterogeneity among bacterial species while yeast microbiota was dominated by Aureobasidium pullulans at DNA level. The obtained results represent a first approach in the understanding of the Planalto de Bolona cheese microbial ecology and consequently may constitute a first step towards the full comprehension of the microbiota of this artisanal cheese produced in unusual environmental conditions in the Cape Verde Islands.




Similar content being viewed by others
References
Abdelgadir WS, Hamad SH, Møller PL, Jakobsen M (2001) Characterisation of the dominant microbiota of Sudanese fermented milk Rob. Int Dairy J 11:63–70
Altschul SF, Madden TL, Shaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ampe F, Ben Omar N, Moizan C, Wacher C, Guyot JP (1999) Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl Environ Microbiol 65:5464–5473
Bankole MO, Okagbue RN (1992) Properties of nono, a Nigerian fermented milk food. Ecol Food Nutr 27:145–149
Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM (2001) Recent advances in cheese microbiology. Int Dairy J 11:259–274
Beukes EM, Bester BH, Mostert JF (2001) The microbiology of South African traditional fermented milks. Int J Food Microbiol 63:189–197
Cocolin L, Bisson LF, Mills DA (2000) Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol Lett 189:81–87
Cocolin L, Manzano M, Aggio D, Cantoni C, Comi G (2001a) A novel polymerase chain reaction (PCR)-denaturing gradient gel electrophoresis (DGGE) for the identification of Micrococcaceae strains involved in meat fermentations. Its application to naturally fermented Italian sausages. Meat Sci 57:59–64
Cocolin L, Manzano M, Cantoni C, Comi G (2001b) Denaturing gradient gel electrophoresis analysis of the 16 S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl Environ Microbiol 67:5113–5121
Cogan TM, Barbosa M, Beuvier E, Bianchi-Salvadori B, Cocconcelli PS, Fernandes I, Gomez J, Gomez R, Kalantzpoulos G, Ledda A, Medina M, Rea MC, Rodriguez E (1997) Characterisation of the lactic acid bacteria in artisanal dairy products. J Dairy Res 64:409–421
Coppola S, Blaiotta G, Ercolini D (2008) Dairy products. In: Cocolin L, Ercolini D (eds) Molecular techniques in the microbial ecology of fermented foods. Springer, New York, pp 31–90
Corbo MR, Lanciotti R, Albenzio M, Sinigaglia M (2001) Occurrence and characterization of yeasts isolated from milks and dairy products of Apulia region. Int J Food Microbiol 69:147–152
Dolci P, Alessandria V, Rantsiou K, Rolle L, Zeppa G, Cocolin L (2008a) Microbial dynamics of Castelmagno PDO, a traditional Italian cheese, with a focus on lactic acid bacteria ecology. Int J Food Microbiol 22:302–311
Dolci P, Alessandria V, Zeppa G, Rantsiou K, Cocolin L (2008b) Microbiological characterization of artisanal Raschera PDO cheese: analysis of its indigenous lactic acid bacteria. Food Microbiol 25:392–399
Ercolini D (2004) PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Meth 56(3):297–314
Ercolini D, Hill PJ, Dodd CER (2003) Bacterial community structure and location in Stilton cheese. Appl Environ Microbiol 69(6):3540–3548
Fleet GH (1990) Yeasts in dairy products. J Appl Bacteriol 68:199–211
Fox PF, McSweeney PLH, Cogan TM, Guinee TP (2004) Cheese: Chemistry, Physics and Microbiology, vol 1, 3rd edn. Elsevier Ltd, London
Gadaga TH, Mutukumira AN, Narvhus JA (2000) Enumeration and identification of yeasts isolated from Zimbabwean traditional fermented milk. Int Dairy J 10:459–466
Garcia Fontan MC, Franco I, Prieto B, Tornadijo ME, Carballo J (2001) Microbiological changes in San Simon cheese throughout ripening and its relationship with physico-chemical parameters. Food Microbiol 18:25–33
Jacques N, Casaregola S (2008) Safety assessment of dairy microorganisms: the hemiascomycetous yeasts. Int J Food Microbiol 126:321–326
Jespensen L (2003) Occurrence and taxonomic characteristics of strains of Saccharomyces cerevisiae predominant in African indigenous fermented food and beverages. FEMS Yeast Res 3:191–200
Klijn N, Weerkamp AH, De Vos WM (1991) Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16 S rRNA and specific DNA probes. Appl Environ Microbiol 57:3390–3393
Kurtzman CP, Robnett CJ (1997) Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26 S) ribosomal DNA gene. J Clin Microbiol 35:1216–1223
Lopandic K, Zelger S, Banszky LK, Eliskases-Lechner F, Prillinger H (2006) Identification of yeasts associated with milk products using traditional and molecular techniques. Food Microbiol 23:341–350
Lopez I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, Marshall M, VanderGheynst J, Mills D (2003) Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:6801–6807
Pereira-Dias S, Potes ME, Marinho A, Malfeito-Ferreira M, Loureiro V (2000) Characterisation of yeast flora isolated from an artisanal Portuguese ewes’ cheese. Int J Food Microbiol 60:55–63
Prillinger H, Molnar O, Eliskases-Lechner F, Lopandic K (1999) Phenotypic and genotypic identification of yeasts from cheese. Antonie Van Leeuwenhoek 75:267–283
Randazzo CL, Torriani S, Akkermans ADL, De Vos WM, Vaughan EE (2002) Diversity, dynamics, and activity of bacterial communities during production of an artisanal sicilian cheese as evaluated by 16S rRNA analysis. Appl Environ Microbiol 68:1882–1892
Rantsiou K, Cocolin L (2006) New developments in the study of the microbiota of naturally fermented sausages as determined by molecular methods: a review. Int J Food Microbiol 108:255–267
Rantsiou K, Urso R, Dolci P, Comi G, Cocolin L (2008) Microflora of Feta cheese from four Greek manufacturers. Int J Food Microbiol 126:36–42
Roostita R, Fleet GH (1996) Growth of yeasts in milk and associated changes to milk composition. Int J Food Microbiol 31:215–219
Sheffield VC, Cox DR, Lerman LS, Myers RM (1989) Attachment of a 40-base pairs G + C rich sequence (GC clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Protoc Natl Acad Sci 86:297–303
Acknowledgments
Authors express their gratitude to Prof. Giuseppe Quaranta, Department of Animal Pathology, Faculty of Veterinary Medicine, University of Turin, from the collection of the samples in the Cape Verde Islands.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alessandria, V., Dolci, P., Rantsiou, K. et al. Microbiota of the Planalto de Bolona: an artisanal cheese produced in uncommon environmental conditions in the Cape Verde Islands. World J Microbiol Biotechnol 26, 2211–2221 (2010). https://doi.org/10.1007/s11274-010-0406-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-010-0406-7