Abstract
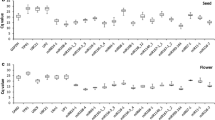
Expression profiling of miRNAs has the ability to reveal the essence of somatic embryogenesis (SE). qRT-PCR is one of the most commonly used techniques for dynamic miRNA detection but requires optimal reference genes for data reliability. This is the first report on reference gene validation for miRNA expression normalization in Lilium (Lilium pumilum DC. Fisch. and Lilium davidii var. unicolor). In this study, seventeen miRNAs together with two snRNAs (U4, U6), one rRNA (5S rRNA) and three protein-coding genes (FP, ACT, GAPDH) were selected as reference candidates, and their expression stability was validated by qRT-PCR among eleven develo** SE cultures in two lilies. Four normalization algorithms, including geNorm, BestKeeper, NormFinder and RefFinder, were also used to evaluate the stability of the reference candidates. For Lilium pumilum DC. Fisch., lpu-miR159a was the optimal reference gene during SE, followed by lpu-miR408b, while U6 was the least stable reference candidate. For Lilium davidii var. unicolor, FP presented greater stability than did half of the miRNA candidates, but the best reference gene was lda-miR162, followed by lda-miR159a. Further analysis of the expression level of miR156 and miR529 was used to evaluate the validity of the reference genes in both lilies. In general, miRNAs are superior to common protein-coding genes and snRNAs / rRNAs as reference genes for miRNA expression normalization during Lilium SE, and the most suitable reference miRNA is different between two species in the same Lilium genus. This is a pioneer study using suitable miRNAs as reference genes in Lilium and constitutes a small but essential step for the further exploration of miRNA function in Lilium, thus offering valuable references for other plants.



Similar content being viewed by others
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. doi:10.1158/0008-5472.CAN-04-0496
Belli Kullan J, Lopes Paim Pinto D, Bertolini E, Fasoli M, Zenoni S, Tornielli GB, Pezzotti M, Meyers BC, Farina L, Pè ME, Mica E (2015) miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genomics 16:393. doi:10.1186/s12864-015-1610-5
Bi W, Yin Z, Guo L, Chen L, Pan C, Wang Q (2015) Plant regeneration from shoot regrowth and de novo embryo-like structures from cryopreserved shoot tips of Lilium spp. In Vitro Cell Dev Biol Plant 51:390–398. doi:10.1007/s11627-015-9696-7
Borges A, Tsai SM, Caldas DGG (2012) Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep 31:827–838. doi:10.1007/s00299-011-1204-x
Borowski JM, Galli V, Messias Rda S, Perin EC, Buss JH, dos Anjos e Silva SD, Rombaldi CV (2014) Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 239:1187–1200. doi:10.1007/s00425-014-2041-2
Candar-Cakir B, Arican E, Zhang B (2016) Small RNA and degradome deep sequencing reveals drought-and tissue-specific microRNAs and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J 14:1727–1746. doi:10.1111/pbi.12533
Cassol D, Cruz FP, Espindola K, Mangeon A, Müller C, Loureiro ME, Corrêa RL, Sachetto-Martins G (2016) Identification of reference genes for quantitative RT-PCR analysis of microRNAs and mRNAs in castor bean (Ricinus communis L.) under drought stress. Plant Physiol Biochem 106:101–107. doi:10.1016/j.plaphy.2016.02.024
Chandna R, Augustine R, Bisht NC (2012) Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS ONE 7:e36918. doi:10.1371/journal.pone.0036918
Chávez-Hernández EC, Alejandri-Ramírez ND, Juárez-González VT, Dinkova TD (2015) Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front Plant Sci 6:555. doi:10.3389/fpls.2015.00555
Chi X, Hu R, Yang Q, Zhang X, Pan L, Chen N, Chen M, Yang Z, Wang T, He Y, Yu S (2012) Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genomics 287:167–176. doi:10.1007/s00438-011-0665-5
Chuck G, Whipple C, Jackson D, Hake S (2010) The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137:1243–1250. doi:10.1242/dev.048348
Ding Y, Chen Z, Zhu C (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62:3563–3573. doi:10.1093/jxb/err046
Fehér A (2015) Somatic embryogenesis—stress-induced remodeling of plant cell fate. Biochim Biophys Acta 1849:385–402. doi:10.1016/j.bbagrm.2014.07.005
Feng H, Huang X, Zhang Q, Wei G, Wang X, Kang Z (2012) Selection of suitable inner reference genes for relative quantification expression of microRNA in wheat. Plant Physiol Biochem 51:116–122. doi:10.1016/j.plaphy.2011.10.010
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618. doi:10.1111/j.1467-7652.2008.00346.x
Haensch KT (1996) Plant regeneration through somatic embryogenesis in different genotypes of Lilium hybrids. Gartenbauwissenschaft 61:214–218
Han H, Guo C (2009) The tissue culture method of Lanzhou Lily. J Tian** Normal Univ (Nat Sci Ed) 29:62–65
He H, Liu D, Zhang N, Zheng W, Han Q, Ji B, Ge F, Chen C (2014) The PR10 gene family is highly expressed in Lilium regale Wilson during Fusarium oxysporum f. sp. lilii infection. Genes Genomics 36:497–507. doi:10.1007/s13258-014-0185-x
Jia L, Zhang D, Qi X, Ma B, **ang Z, He N (2014) Identification of the conserved and novel miRNAs in mulberry by high-throughput sequencing. PLoS ONE 9:e104409. doi:10.1371/journal.pone.0104409
** L, Zhang Y, Yan L, Guo Y, Niu L (2012) Phenolic compounds and antioxidant activity of bulb extracts of six Lilium species native to China. Molecules 17:9361–9378. doi:10.3390/molecules17089361
Kou SJ, Wu XM, Liu Z, Liu YL, Xu Q, Guo WW (2012) Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep 31:2151–2163. doi:10.1007/s00299-012-1325-x
Koul A, Yogindran S, Sharma D, Kaul S, Rajam MV, Dhar MK (2016) Carotenoid profiling, in silico analysis and transcript profiling of miRNAs targeting carotenoid biosynthetic pathway genes in different developmental tissues of tomato. Plant Physiol Biochem 108:412–421. doi:10.1016/j.plaphy.2016.08.001
Kulcheski FR, Marcelino-Guimaraes FC, Nepomuceno AL, Abdelnoor RV, Margis R (2010) The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal Biochem 406:185–192. doi:10.1016/j.ab.2010.07.020
Li XY, Wang CX, Sun HM, Li TL (2011) Establishment of the total RNA extraction system for lily bulbs with abundant polysaccharides. Afr J Biotechnol 10:17907–17915
Li Q, Fan CM, Zhang XM, Fu YF (2012a) Validation of reference genes for real-time quantitative PCR normalization in soybean developmental and germinating seeds. Plant Cell Rep 31:1789–1798. doi:10.1007/s00299-012-1282-4
Li T, Chen J, Qiu S, Zhang Y, Wang P, Yang L, Lu Y, Shi J (2012b) Deep sequencing and microarray hybridization identify conserved and species-specific microRNAs during somatic embryogenesis in hybrid yellow poplar. PLoS ONE 7:e43451. doi:10.1371/journal.pone.0043451
Li H, Mao W, Liu W, Dai H, Liu Y, Ma Y, Zhang Z (2013) Deep sequencing discovery of novel and conserved microRNAs in wild type and a white-flesh mutant strawberry. Planta 238:695–713. doi:10.1007/s00425-013-1917-x
Li W, Zhang S, Han S, Wu T, Zhang J, Qi L (2014) The post-transcriptional regulation of LaSCL6 by miR171 during maintenance of embryogenic potential in Larix kaempferi (lamb.) Carr. Tree Genet Genomes 10:223–229. doi:10.1007/s11295-013-0668-y
Li X, Cheng J, Zhang J, Teixeira da Silva JA, Wang C, Sun H (2015) Validation of reference genes for accurate normalization of gene expression in Lilium davidii var. unicolor for real time quantitative PCR. PLoS ONE 10:e0141323. doi:10.1371/journal.pone.0141323
Lin YL, Lai ZX (2010) Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci 178:359–365. doi:10.1016/j.plantsci.2010.02.005
Lin YL, Lai ZX (2013) Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol Biochem 66:20–25. doi:10.1016/j.plaphy.2013.02.002
Lin Y, Lai Z, Tian Q, Lin L, Lai R, Yang M, Zhang D, Chen Y, Zhang Z (2015) Endogenous target mimics down-regulate miR160 mediation of ARF10, -16, and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front Plant Sci 6:956. doi:10.3389/fpls.2015.00956
Liu Y, Wang L, Chen D, Wu X, Huang D, Chen L, Li L, Deng X, Xu Q (2014) Genome-wide comparison of microRNAs and their targeted transcripts among leaf, flower and fruit of sweet orange. BMC Genomics 15:695. doi:10.1186/1471-2164-15-695
Lugassi-Ben Hamo M, Martin CV, Zaccai M (2015) Characterization of expressed sequence tags from Lilium longiflorumin vernalized and non-vernalized bulbs. J Plant Physiol 173:72–81. doi:10.1016/j.jplph.2014.09.015
Machado RD, Christoff AP, Loss-Morais G, Margis-Pinheiro M, Margis R, Körbes AP (2015) Comprehensive selection of reference genes for quantitative gene expression analysis during seed development in Brassica napus. Plant Cell Rep 34:1139–1149. doi:10.1007/s00299-015-1773-1
Maroufi A, Van Bockstaele E, De Loose M (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15. doi:10.1186/1471-2199-11-15
Micheli M, Standard A (2016) From somatic embryo to synthetic seed in Citrus spp through the encapsulation technology. In: Germana MA, Lambardi M (eds) In vitro embryogenesis in higher plants. Springer, New York, pp 515–522
Nakano M, Sakakibara T, Suzuki S, Saito H, Sakakiba T (2000) Decrease in the regeneration potential of long-term cell suspension cultures of Lilium formosanum Wallace and its restoration by the auxin transport inhibitor, 2,3,5-triiodobenzoic acid. Plant Sci 158:129–137. doi:10.1016/S0168-9452(00)00313-7
Nakhooda M, Mandiri E (2016) Using synergistic exogenous phytohormones to enhance somatic embryogenesis from leaf explants of a eucalyptus grandis clone. Revista Bellas Artes 6:35–39
Nhut DT, Hanh NTM, Tuan PQ, Nguyet LTM, Tram NTH, Chinh NC, Nguyen NH, Vinh DN (2006) Liquid culture as a positive condition to induce and enhance quality and quantity of somatic embryogenesis of Lilium longiflorum. Sci Hortic 110:93–97. doi:10.1016/j.scienta.2006.05.015
Ooi S, Choo C, Ishak Z, Ong-Abdullah M (2012) A candidate auxin-responsive expression marker gene, EgIAA9, for somatic embryogenesis in oil palm (Elaeis guineensis Jacq.). Plant Cell Tissue Organ Cult 110:201–212. doi:10.1007/s11240-012-0143-8
Pitschmann A, Purevsuren S, Obmann A, Natsagdorj D, Gunbilig D, Narantuya S, Kletter C, Glasl S (2013) Traditional Mongolian medicine: history and status quo. Phytochem Rev 12:943–959. doi:10.1007/s11101-013-9321-5
Pullman GS, Olson K, Fischer T, Egertsdotter U, Frampton J, Bucalo K (2016) Fraser fir somatic embryogenesis: high frequency initiation, maintenance, embryo development, germination and cryopreservation. New For 47:453–480. doi:10.1007/s11056-016-9525-9
Qi S, Yang L, Wen X, Hong Y, Song X, Zhang M, Dai S (2016) Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front Plant Sci 7:287. doi:10.3389/fpls.2016.00287
Qiao M, Zhao Z, Song Y, Liu Z, Cao L, Yu Y, Li S, **ang F (2012) Proper regeneration from in vitro cultured Arabidopsis thaliana requires the microRNA-directed action of an auxin response factor. Plant J 71:14–22. doi:10.1111/j.1365-313X.2012.04944.x
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349. doi:10.1007/s00425-008-0706-4
Shen Y, Jiang Z, Lu S, Lin H, Gao S, Peng H, Yuan G, Liu L, Zhang Z, Zhao M, Rong T, Pan G (2013) Combined small RNA and degradome sequencing reveals microRNA regulation during immature maize embryo dedifferentiation. Biochem Biophys Res Commun 441:425–430. doi:10.1016/j.bbrc.2013.10.113
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39:519–525. doi:10.2144/000112010
Silva AT, Barduche D, do Livramento KG, Ligterink W, Paiva LV (2014) Characterization of a putative Serk-Like ortholog in embryogenic cell suspension cultures of Coffea arabica L. Plant Mol Biol Rep 32:176–184. doi:10.1007/s11105-013-0632-x
Song H, Zhang X, Shi C, Wang S, Wu A, Wei C (2016) Selection and verification of candidate reference genes for mature microRNA expression by quantitative RT-PCR in the tea plant (Camellia sinensis). Genes 7:25. doi:10.3390/genes7060025
Sun SK, Yang NN, Chen LJ, Irfan M, Zhao XH, Li TL (2015) Characterization of LpGPAT gene in Lilium pensylvanicum and response to cold stress. Biomed Res Int 2015:792819. doi:10.1155/2015/792819
Sun H, Li F, Ruan Q, Zhong X (2016) Identification and validation of reference genes for quantitative real-time PCR studies in Hedera helix L. Plant Physiol Biochem 108:286–294. doi:10.1016/j.plaphy.2016.07.022
Tang N, Mo G, van Tuyl JM, Arens P, Liu JJ, Tang DC (2014) Genetic diversity and structure of Lilium pumilum DC. in southeast of Qinghai–Tibet plateau. Plant Syst Evol 300:1453–1464. doi:10.1007/s00606-013-0973-9
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. doi:10.1186/1746-4811-3-12
Vega-Bartol JJD, Santos R, Simões M, Miguel C (2011) Evaluation of reference genes for quantitative PCR analysis during somatic embryogenesis in conifers. BMC Proc 5:O44
Wang T, Lu J, Xu Z, Yang W, Wang J, Cheng T, Zhang Q (2014) Selection of suitable reference genes for miRNA expression normalization by qRT-PCR during flower development and different genotypes of Prunus mume. Sci Hortic 169:130–137. doi:10.1016/j.scienta.2014.02.006
Wang Y, Lan Q, Zhao X, Xu W, Li F, Wang Q, Chen R (2016) Comparative profiling of microRNA expression in soybean seeds from genetically modified plants and their near-isogenic parental lines. PLoS ONE 11:e0155896. doi:10.1371/journal.pone.0155896
Wu XM, Liu MY, Ge XX, Xu Q, Guo WW (2011) Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 233:495–505. doi:10.1007/s00425-010-1312-9
Wu XM, Kou SJ, Liu YL, Fang YN, Xu Q, Guo WW (2015) Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA-and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol J 13:383–394. doi:10.1111/pbi.12317
Yan J, Yuan F, Long G, Qin L, Deng Z (2012) Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol Biol Rep 39:1831–1838. doi:10.1007/s11033-011-0925-9
Yang L, Lei B, Shen HL, Li YH (2013a) Somatic embryogenesis and plantlet regeneration from mature zygotic embryos of Manchurian ash (Fraxinus mandshurica Rupr.). Plant Cell Tissue Organ Cult 115:115–125. doi:10.1007/s11240-013-0345-8
Yang X, Wang L, Yuan D, Lindsey K, Zhang X (2013b) Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J Exp Bot 64:1521–1536. doi:10.1093/jxb/ert013
Yin ZF, Chen L, Bi WL, Wang QC, Volk GM (2014) Somatic embryogenesis and organogenesis from cryopreserved shoot tips of Lilium oriental hybrid ‘Siberia’. Acta Hortic 1039:193–200
Zhang LF, Li WF, Han SY, Yang WH, Qi LW (2013) cDNA cloning, genomic organization and expression analysis during somatic embryogenesis of the translationally controlled tumor protein (TCTP) gene from Japanese larch (Larix leptolepis). Gene 529:150–158. doi:10.1016/j.gene.2013.07.076
Zhang L, Li W, Xu H, Qi L, Han S (2014) Cloning and characterization of four differentially expressed cDNAs encoding NFYA homologs involved in responses to ABA during somatic embryogenesis in Japanese larch (Larix leptolepis). Plant Cell Tissue Organ Cult 117:293–304. doi:10.1007/s11240-014-0440-5
Zhang J, Gai M, Li X, Li T, Sun H (2016) Somatic embryogenesis and direct as well as indirect organogenesis in Lilium pumilum DC. Fisch., an endangered ornamental and medicinal plant. Biosci Biotechnol Biochem 80:1898–1906. doi:10.1080/09168451.2016.1194178
Acknowledgements
This project was supported by the National Natural Science Foundation of China [grant number 31471897; 31672179] and the Program for Excellent Talents in the University of Liaoning province, China [grant number LR2013029].
Author contributions
JZ and HMS conceived and designed the experiment. MZG and BYX contributed reagents and materials. CXW designed the primers for qRT-PCR. JZ, MZG, BYX and NNJ performed RNA extraction and qRT-PCR. JZ and JXW analyzed the data. JZ and HMS wrote and revised the manuscript. All of the authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in the submission of this manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Gai, M., Xue, B. et al. The use of miRNAs as reference genes for miRNA expression normalization during Lilium somatic embryogenesis by real-time reverse transcription PCR analysis. Plant Cell Tiss Organ Cult 129, 105–118 (2017). https://doi.org/10.1007/s11240-016-1160-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-016-1160-9