Abstract
Key message
CmCAD2 and CmCAD3 function more positively than CmCAD1 in oriental melon for lignin synthesis which is important to ensure internal water status and thus for drought tolerance.
Abstract
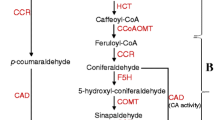
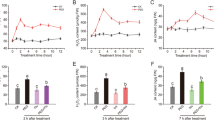
Well-lignification may be the guarantee of efficient axial water transport and barrier of lateral water flow in oriental melon tolerating drought stress, however remains to be verified. As an important enzyme in monolignol synthesis pathway, five cinnamyl alcohol dehydrogenase (CAD) genes were generally induced in melon seedlings by drought. Here we further revealed the roles of CmCAD1, 2, and 3 in lignin synthesis and for drought tolerance. Results found that overexpressing CmCAD2 or 3 strongly recovered CAD activities, lignin synthesis and composition in Arabidopsis cadc cadd, whose lignin synthesis is disrupted, while CmCAD1 functioned modestly. In melon seedlings, silenced CmCAD2 and 3 individually or collectively decreased CAD activities and lignin depositions drastically, resulting in dwarfed phenotypes. Reduced lignin, mainly composed by guaiacyl units catalyzed by CmCAD3, is mainly due to the limited lignification in tracheary elements and development of Casparion strip. While CmCAD1 and 2 exhibited catalysis to p-coumaraldehyde and sinapaldehyde, respectively. Compared with CmCAD1, drought treatments revealed higher sensitivity of CmCAD2 and/or 3 silenced melon seedlings, accompanying with lower relative water contents, water potentials and relatively higher total soluble sugar contents. Slightly up-regulated expressions of aquaporin genes together with limited lignification might imply higher lateral water loss in stems of silenced lines. In Arabidopsis, CmCAD2 and 3 transgenic lines enhanced cadc cadd drought tolerance through recovering lignin synthesis and root development, accompanying with decreased electrolyte leakage ratios and increased RWCs, thus improved survival rates. Briefly, lignin synthesized by CmCAD2 and 3 functions importantly for drought tolerance in melon.










Similar content being viewed by others
References
Alexandersson E, Fraysse L, Sjövall-Larsen S, Gustavsson S, Fellert M, Karlsson M et al (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59:469–484
Al-Hakimi AMA (2006) Counteraction of drought stress on soybean plants by seed soaking in salicylic acid. Int J Bot 2:421–426
Alvarez S, Marsh EL, Schroeder SG, Schachtman DP (2008) Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ 31:325–340
Anderson NA, Tobimatsu Y, Ciesielski PN, **menes E, Ralph J, Donohoe BS et al (2015) Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27:2195–2209
Bang SW, Lee DK, Jung H, Chung PJ, Kim YS, Choi YD et al (2019) Overexpression of OsTF1L, a rice HD-Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol J 17:118–131
Barakat A, Bagniewska-Zadworna A, Frost CJ, Carlson JE (2010) Phylogeny and expression profiling of CAD and CAD-like genes in hybrid Populus (P. deltoides × P. nigra): evidence from herbivore damage for subfunctionalization and functional divergence. BMC Plant Biol. 10:100
Black RA, Rosen AA, Adams SL (1953) The chromatographic separation of hardwood extractive components giving color reactions with phloroglucinol. J Am Chem Soc 75:5344–5346
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–363
Boudet AM (2000) Lignin and lignification: selected issues. Plant Physiol Biochem 38:81–96
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chabannes M, Barakate A, Lapierre C, Marita JM, Ralph J, Pean M et al (2001) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J 28:257–270
Chapple C, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4:1413–1424
Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X et al (2013) A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant 6:1769–1780
Chen W, VanOpdorp N, Fitzl D, Tewari J, Friedemann P, Greene T et al (2012) Transposon insertion in a cinnamyl alcohol dehydrogenase gene is responsible for a brown midrib1 mutation in maize. Plant Mol Biol 80:289–297
Cui GB, Zhao YF, Zhang JL, Chao MN, **e KL, Zhang C et al (2019) Proteomic analysis of the similarities and differences of soil drought and polyethylene glycol stress responses in wheat (Triticum aestivum L.). Plant Mol Biol 100:391–410
Dong HP, Yu HQ, Bao ZL, Guo XJ, Peng JL, Yao Z et al (2005) The ABI2-dependent abscisic acid signalling controls HrpN-induced drought tolerance in Arabidopsis. Planta 221:313–327
Du Y, Zhao Q, Chen L, Yao X, Zhang W, Zhang B et al (2020) Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol Biochem 146:1–12
Eudes A, Pollet B, Sibout R, Do CT, Séguin A, Lapierre C, Jouanin L (2006) Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225:23–39
Fan L, Linker R, Gepstein S, Tanimoto E, Yamamoto R, Neumann PM (2006) Progressive inhibition by water deficit of cell wall extensibility and growth along the elongation zone of maize roots is related to increased lignin metabolism and progressive stelar accumulation of wall phenolics. Plant Physiol 140:603–612
Fornalé S, Capellades M, Encina A, Wang K, Irar S, Lapierre C et al (2012) Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol Plant 5:817–830
Fornalé S, Rencoret J, García-Calvo L, Encina A, Rigau J, Gutiérrez A et al (2017) Changes in cell wall polymers and degradability in maize mutants lacking 3′- and 5′-O-methyltransferases involved in lignin biosynthesis. Plant Cell Physiol 58:240–255
Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C et al (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16:1446–1465
Hose E, Clarkson DT, Steudle E, Schreiber L, Hartung W (2001) The exodermis: a variable apoplastic barrier. J Exp Bot 52:2245–2264
Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD et al (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17:808
** YZ, Zhang C, Liu W, Qi HY, Chen H, Cao SX (2014) The cinnamyl alcohol dehydrogenase gene family in melon (Cucumis melo L.): Bioinformatic analysis and expression patterns. PLoS ONE 9:e101730
Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L et al (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126:1358–1369
Jubany-Marí T, Munné-Bosch S, López-Carbonell M, Alegre L (2009) Hydrogen peroxide is involved in the acclimation of the Mediterranean shrub, Cistus albidus L., to summer drought. J Exp Bot 60:107–120
Kim SJ, Kim MR, Bedgar DL, Moinuddin SGA, Cardenas CL, Davin LB et al (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101:1455–1460
Kuromori T, Seo M, Shinozaki K (2018) ABA transport and plant water stress responses. Trends Plant Sci 23:513–522
Lee BR, Muneer S, Jung WJ, Avice JC, Ourry A, Kim TH (2012) Mycorrhizal colonization alleviates drought-induced oxidative damage and lignification in the leaves of drought-stressed perennial ryegrass (Lolium perenne). Physiol Plant 145:440–449
Lee DK, Yoon S, Kim YS, Kim JK (2017) Rice OsERF71-mediated root modification affects shoot drought tolerance. Plant Signal Behav 12:e1268311
Li J, Yang P, Yang Q, Gong X, Ma H, Dang K et al (2019) Analysis of flavonoid metabolites in buckwheat leaves using UPLC-ESI-MS/MS. Molecules 24:1310
Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22:1620–1632
Li Z, Peng Y, Ma X (2013) Different response on drought tolerance and post-drought recovery between the small-leafed and the large-leafed white clover (Trifolium repens L.) associated with antioxidative enzyme protection and lignin metabolism. Acta Physiol Plant 35:213–222
Lin SY, Dence CW (1992) Methods in lignin chemistry. Springer, Berlin
Liu W, ** YZ, Li MM, Dong LJ, Guo D, Lu C, Qi HY (2018) Analysis of CmCADs and three lignifying enzymes in oriental melon (‘CaiHong7’) seedlings in response to three abiotic stresses. Sci Hortic-Amst 237:257–268
Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31:777–786
Liu YL, Sun W, Zeng SH, Huang WJ, Liu D, Hu WM et al (2014) Virus-induced gene silencing in two novel functional plants, Lycium barbarum L. and Lycium ruthenicum Murr. Sci Hortic-Amst 170:267–274
Mir Derikvand M, Sierra JB, Ruel K, Pollet B, Do CT, Thévenin J et al (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227:943–956
Moschen S, Di Rienzo JA, Higgins J, Tohge T, Watanabe M, González S et al (2017) Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol Biol 94:549–564
Moura-Sobczak J, Souza U, Mazzafera P (2011) Drought stress and changes in the lignin content and composition in Eucalyptus. BMC Proc 5:P103
Moura JCMS, Bonine CAV, De Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52:360–376
Muro-Villanueva F, Mao X, Chapple C (2019) Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr Opin Biotechnol 56:202–208
Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109:10101–10106
Park SC, Kim YH, Jeong JC, Kim CY, Lee HS, Bang JW et al (2011) Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta 233:621–634
Pedersen JF, Vogel KP, Funnell DL (2005) Impact of reduced lignin on plant fitness. Crop Sci 45:812–819
Reddy MSS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci USA 102:16573–16578
Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60:771–782
Schreiber L (1996) Chemical composition of Casparian strips isolated from Clivia miniata Reg. roots: evidence for lignin. Planta 199:596–601
Shadle G, Chen F, Srinivasa Reddy MS, Jackson L, Nakashima J, Dixon RA (2007) Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68:1521–1529
Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17:2059–2076
Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F et al (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol 132:848–860
Song L, Prince S, Valliyodan B, Joshi T, Maldonado dos Santos JV, Wang JJ et al (2016) Genome-wide transcriptome analysis of soybean primary root under varying water-deficit conditions. BMC Genomics 17:57
Srivastava S, Vishwakarma RK, Arafat YA, Gupta SK, Khan BM (2015) Abiotic stress induces change in Cinnamoyl CoA Reductase (CCR) protein abundance and lignin deposition in develo** seedlings of Leucaena leucocephala. Physiol Mol Biol Plants 21:197–205
Tobias CM, Chow EK (2005) Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220:678–688
Van Acker R, Déjardin A, Desmet S, Hoengenaert L, Vanholme R, Morreel K et al (2017) Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol 175:1018–1039
Vincent D, Lapierre C, Pollet B, Cornic G, Negroni L, Zivy M (2005) Water deficits affect caffeate o-methyltransferase, lignification, and related enzymes in maize leaves A proteomic investigation. Plant Physiol 137:949–960
Voelker SL, Lachenbruch B, Meinzer FC, Kitin P, Strauss SH (2011) Transgenic poplars with reduced lignin show impaired xylem conductivity, growth efficiency and survival. Plant Cell Environ 34:655–668
Wagner A, Tobimatsu Y, Goeminne G, Phillips L, Flint H, Steward D et al (2013) Suppression of CCR impacts metabolite profile and cell wall composition in Pinus radiata tracheary elements. Plant Mol Biol 81:105–117
Wang X, Cai H, Li Y, Zhu YM, Ji W, Bai X et al (2015) Ectopic overexpression of a novel Glycine soja stress-induced plasma membrane intrinsic protein increases sensitivity to salt and dehydration in transgenic Arabidopsis thaliana plants. J Plant Res 128:103–113
Yamaguchi M, Valliyodan B, Zhang J, Lenoble ME, Yu O, Rogers EE et al (2010) Regulation of growth response to water stress in the soybean primary root. I. Proteomic analysis reveals region-specific regulation of phenylpropanoid metabolism and control of free iron in the elongation zone. Plant Cell Environ 33:223–243
Yan JW, Aznar A, Chalvin C, Birdseye DS, Baidoo EEK, Eudes A et al (2018) Increased drought tolerance in plants engineered for low lignin and low xylan content. Biotechnol Biofuels 11:195
Yoshimura K, Masuda A, Kuwano M, Yokota A, Akashi K (2008) Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol 49:226–241
Zhang LB, Wang G, Chang JM, Liu J, Cai JH, Rao XW et al (2010) Effects of 1-MCP and ethylene on expression of three CAD genes and lignification in stems of harvested Tsai Tai (Brassica chinensis). Food Chem 123:32–40
Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646
Zhao Q, Tobimatsu Y, Zhou R, Pattathil S, Gallego-Giraldo L, Fu CX et al (2013) Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc Natl Acad Sci USA 110:13660–13665
Acknowledgements
We are quite grateful to Prof. Clint Chapple (Purdue University, West Lafayette, Indiana) for kindly gifting us the Arabidopsis cadc cadd double mutant. We would also like to thanks Prof. Tao Xu and Prof. Feng Wang for their kindly guides.
Funding
This work was supported by the Agriculture Research System of China (CARS-25).
Author information
Authors and Affiliations
Contributions
WL, YJ and HYQ conceived and designed the experiments. WL and CHW obtained the materials of VIGS melon seedlings and Arabidopsis transgenic lines, WL and YJ performed the related experiments. YZJ helped to do the phylogeny analysis. WL and QJX carried out the mannitol treatment. WL, ML and THL carried out the TEM experiments. WL and LLZ analyzed the data. WL and YJ completed the manuscript and HYQ improved it.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Accession numbers: Sequence data can be achieved in the Melonomics (https://www.melonomics.net/): CmCAD1 (MELO3C019548), CmCAD2 (MELO3C018492), CmCAD3 (MELO3C003735), CmCAD4 (MELO3C005809), CmCAD5 (MELO3C023272), CmPIP2;1 (MELO3C025772), CmPIP2;2 (MELO3C013347), CmPIP2;3 (MELO3C019794), CmPIP2;6 (MELO3C014239), CmTIP1;1 (MELO3C024483), CmTIP2;2 (MELO3C005526), CmTIP4;1 (MELO3C011146), CmTIP5;1 (MELO3C005441), CmNIP1;1 (MELO3C007188), CmNIP2;1 (MELO3C009870), CmNIP3;2 (MELO3C005817), CmNIP4;1 (MELO3C006559).
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2020_1018_MOESM1_ESM.tif
Semi-quantitative RT-PCR (A) and qRT-PCR (B) analysis of the expression patterns of five CmCAD genes in leaf, stem and root. Supplementary file1 (TIF 1634 kb)
11103_2020_1018_MOESM2_ESM.tif
Phylogeny analysis of the deduced amino acid sequences of 5 CmCADs with 16 PoptrCADs from hybrid Populus (Barakat et al., 2010), 9 AtCADs from Arabidopsis thaliana (Sibout et al., 2005; Eudes et al., 2006) and 11 OsCADs from rice (Oryza sativa) (Tobias and Chow, 2005). CmCADs are highlighted with red dots. Supplementary file2 (TIF 6633 kb)
11103_2020_1018_MOESM3_ESM.tif
qRT-PCR detection of CmCAD1, CmCAD2 and CmCAD3 expressions in the 8-week-old CmCADs transgenic Arabidopsis lines. Supplementary file3 (TIF 3355 kb)
11103_2020_1018_MOESM4_ESM.tif
Thickness of vascular tissue and lignified fiber cells. The thickness of vascular tissue and lignified fiber cells how we calculated is indicated in the upper graph. Supplementary file4 (TIF 3749 kb)
11103_2020_1018_MOESM5_ESM.tif
Leaf area (A), internode length (B) and internode diameter (C) of 20-day-old CmCADs silenced melon seedlings. Supplementary file5 (TIF 2857 kb)
11103_2020_1018_MOESM6_ESM.tif
qRT-PCR analysis of silencing efficiency of the 3 target CmCADs as well as CmCAD4 and 5 in stems (A) and leaves (B) of 20-day-old CmCADs silenced melon seedlings. Supplementary file6 (TIF 4558 kb)
11103_2020_1018_MOESM7_ESM.tif
Morphology (A), survival rate (B), root length (C) and electrolyte leakage (D) of WT, cadc cadd and CmCADs transgenic Arabidopsis lines under drought stress simulated by 0, 300 and 400 mmol/L mannitol treatment for 12 days. Bars = 1cm. Supplementary file7 (TIF 6242 kb)
11103_2020_1018_MOESM8_ESM.tif
Multiple alignment of the predicted amino acid sequences of 9 melon PIPs with 13 Arabidopsis thaliana PIPs. Amino acid sequences are aligned by DNAMAN software. Six transmembrane-helix (H1 to H6) are shown by black lines above the sequences. The most highly conserved amino acid sequences (NPA motif) are indicated using green boxes. Residues forming the ar/R selectivity filter and reported to confer glycerol selectivity (P1–P5) are mapped onto the alignment with blue and red boxes, respectively. Supplementary file8 (TIF 8745 kb)
11103_2020_1018_MOESM9_ESM.tif
Multiple alignment of the predicted amino acid sequences of 7 melon TIPs with 10 Arabidopsis thaliana TIPs. Amino acid sequences are aligned by DNAMAN software. Six transmembrane-helix (H1 to H6) are shown by black lines above the sequences. The most highly conserved amino acid sequences (NPA motif) are indicated using green boxes. Residues forming the ar/R selectivity filter and reported to confer glycerol selectivity (P1–P5) are mapped onto the alignment with blue and red boxes, respectively. Supplementary file9 (TIF 6848 kb)
11103_2020_1018_MOESM10_ESM.tif
Multiple alignment of the predicted amino acid sequences of 8 melon NIPs with 9 Arabidopsis thaliana NIPs. Amino acid sequences are aligned by DNAMAN software. Six transmembrane-helix (H1 to H6) are shown by black lines above the sequences. The most highly conserved amino acid sequences (NPA motif) are indicated using green boxes. Residues forming the ar/R selectivity filter and reported to confer glycerol selectivity (P1–P5) are mapped onto the alignment with blue and red boxes, respectively. Supplementary file10 (TIF 7127 kb)
11103_2020_1018_MOESM11_ESM.tif
Multiple alignment of the predicted amino acid sequences of 2 melon SIPs with 3 Arabidopsis thaliana SIPs. Amino acid sequences are aligned by DNAMAN software. Six transmembrane-helix (H1 to H6) are shown by black lines above the sequences. NPA motifs are indicated using green boxes. Residues forming the ar/R selectivity filter and reported to confer glycerol selectivity (P1–P5) are mapped onto the alignment with blue and red boxes, respectively. Supplementary file11 (TIF 2793 kb)
11103_2020_1018_MOESM12_ESM.tif
Phylogenetic relationship between melon AQPs and Arabidopsis thaliana AQPs. The accession numbers corresponding to the sequences used in GenBank are Arabidopsis thaliana AtPIP1;1(At3g61430), ATPIP1;2(At2g45960), AtPIP1;3(At1g01620), AtPIP1;4(At4g00430), AtPIP1;5(At4g23400), AtPIP2;1(At3g53420), AtPIP2;2(At2g37170), AtPIP2;3(At2g37180), AtPIP2;4(At5g60660), AtPIP2;5(At3g54820), AtPIP2;6(At2g39010), AtPIP2;7(At4g35100), AtPIP2;8(At2g16850), AtTIP1;1(At2g36830), AtTIP1;2(At3g26520), AtTIP1;3(At4g01470), AtTIP2;1(At3g16240), AtTIP2;2(At4g17340), AtTIP2;3(At5g47450), AtTIP3;1(At1g73190), AtTIP3;2(At1g17810), AtTIP4;1(At2g25810), AtTIP5;1(At3g47440), AtNIP1;1(At4g19030), AtNIP1;2(At4g18910), AtNIP2;1(At2g34390), AtNIP3;1(At1g31880), AtNIP4;1(At5g37810), AtNIP4;2(At5g37820), AtNIP5;2(At4g10380), AtNIP6;3(At1g80760), AtNIP7;4(At3g06100), AtSIP1;1(At3g04090), AtSIP1;2(At5g18290), AtSIP2;1(At3g56950). The accession numbers corresponding to the sequences used in Melonomics (https://www.melonomics.net/) are Cucumis melo CmPIP1;1(MELO3C005685), CmPIP2;1(MELO3C025772), CmPIP2;2(MELO3C013347), CmPIP2;3(MELO3C019794), CmPIP2;4(MELO3C012429), CmPIP2;5(MELO3C014241), CmPIP2;6(MELO3C014239), CmPIP2;7(MELO3C014240), CmPIP2;8(MELO3C014238), CmTIP1;1(MELO3C024483), CmTIP1;2(MELO3C009377), CmTIP2;1(MELO3C024263), CmTIP2;2(MELO3C005526), CmTIP3;1(MELO3C002183), CmTIP4;1(MELO3C011146), CmTIP5;1(MELO3C005441), CmNIP1;1(MELO3C007188), CmNIP1;2(MELO3C020281), CmNIP2;1(MELO3C009870), CmNIP2;2(MELO3C009871), CmNIP3;1(MELO3C005818), CmNIP3;2(MELO3C005817), CmNIP3;3(MELO3C017831), CmNIP4;1(MELO3C006559), CmSIP1;1(MELO3C008793), CmSIP2;1(MELO3C009719). Numbers above nodes indicate bootstrap support from 1000 pseudoreplicates (in percentage) for selected nodes of interest. Supplementary file12 (TIF 8049 kb)
Rights and permissions
About this article
Cite this article
Liu, W., Jiang, Y., Wang, C. et al. Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance. Plant Mol Biol 103, 689–704 (2020). https://doi.org/10.1007/s11103-020-01018-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01018-7