Abstract
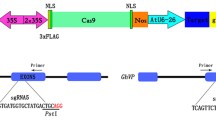
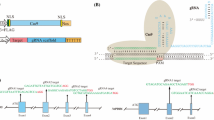
The clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR associated (Cas)9 protein system has emerged as a simple and efficient tool for genome editing in eukaryotic cells. It has been shown to be functional in several crop species, yet there are no reports on the application of this or any other genome editing technologies in the cotton plant. Cotton is an important crop that is grown mainly for its fiber, but its seed also serves as a useful source of edible oil and feed protein. Most of the commercially-grown cotton is tetraploid, thus making it much more difficult to target both sets of homeologous alleles. Therefore, in order to understand the efficacy of the CRISPR/Cas9 system to target a gene within the genome of cotton, we made use of a transgenic cotton line previously generated in our laboratory that had a single copy of the green fluorescent protein (GFP) gene integrated into its genome. We demonstrate, for the first time, the use of this powerful new tool in targeted knockout of a gene residing in the cotton genome. By following the loss of GFP fluorescence, we were able to observe the cells that had undergone targeted mutations as a result of CRISPR/Cas9 activity. In addition, we provide examples of the different types of indels obtained by Cas9-mediated cleavage of the GFP gene, guided by three independent sgRNAs. The results provide useful information that will help us target important native genes in the cotton plant in future.







Similar content being viewed by others
References
Baltes NJ, Voytas DF (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol 33:120–131
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. The Plant Cell 26:151–163
Barrangou R et al (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Baysal C, Bortesi L, Zhu C, Farré G, Schillberg S, Christou P (2016) CRISPR/Cas9 activity in the rice OsBEIIb. Mol Breed 36:1–11. doi:10.1007/s11032-016-0533-4
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Brazelton VA et al (2015) A quick guide to CRISPR sgRNA design tools. GM Crops and Food 6:266–276. doi:10.1080/21645698.2015.1137690
Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297
Cermak T, Starker CG, Voytas DF (2015) Efficient design and assembly of custom TALENs using the golden gate platform. In: Pruett-Miller SM (ed) Chromosomal mutagenesis. Springer New York, New York, NY, pp 133–159
Char SN et al (2017) An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J 15(2):257–268
Chari R, Mali P, Moosburner M, Church GM (2015) Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Meth 12:823–826
Chaudhry B, Yasmin A, Husnain T, Riazuddin S (1999) Mini-scale genomic DNA extraction from cotton. Plant Mol Biol Rep 17:1–7
Cong L, Zhang F (2015) Genome engineering using CRISPR-Cas9 system. In: Pruett-Miller SM (ed) Chromosomal mutagenesis. Springer New York, New York, NY, pp 197–217
Cong L et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Dehairs J, Talebi A, Cherifi Y, Swinnen JV (2016) CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci Rep 6:28973. doi:10.1038/srep28973
Doench JG et al (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotech 32:1262–1267
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109:E2579–E2586
Heigwer F, Kerr G, Boutros M (2014) E-CRISP: fast CRISPR target site identification. Nat Methods 11:122–123
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170
Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S (2015) CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun 467:76–82
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res:1–12. doi:10.1093/nar/gkt780
**ek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–Guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Koonin EV, Makarova KS (2009) CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol Rep 1:95. doi:10.3410/b1-95
Lei Y, Lu L, Liu H-Y, Li S, **ng F, Chen L-L (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7:1494–1496
Li J-F et al (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotech 31:688–691
Li F et al (2015a) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotech 33:524–530
Li J-F, Zhang D, Sheen J (2015b) Targeted plant genome editing via the CRISPR/Cas9 technology. Plant Funct Genomics Methods Protoc 1284:239–255
Li Z et al (2015c) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169:960–970
Liang G, Zhang H, Lou D, Yu D (2016) Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep 6:21451. doi:10.1038/srep21451
Ma X et al (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Mali P, Esvelt KM, Church GM (2013a) Cas9 as a versatile tool for engineering biology. Nat Meth 10:957–963
Mali P et al (2013b) RNA-guided human genome engineering via Cas9. Science 339:823–826
Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotech 31:691–693
Nelson CE, Gersbach CA (2016) Engineering delivery vehicles for genome editing. Annu Rev Chem Biomol Eng 7:637–662. doi:10.1146/annurev-chembioeng-080615-034711
Pan C et al (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6:24765. doi:10.1038/srep24765
Prykhozhij SV, Rajan V, Gaston D, Berman JN (2015) CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PloS ONE 10:e0119372. doi:10.1371/journal.pone.0119372
Rathore KS, Campbell LM, Sherwood S, Nunes E (2015) Cotton (Gossypium hirsutum L.). In: Wang K (ed) Agrobacterium protocols: Vol 2. Springer New York, New York, NY, pp 11–23. doi:10.1007/978-1-4939-1658-0_2
Seth K, Harish (2016) Current status of potential applications of repurposed Cas9 for structural and functional genomics of plants. Biochem Biophys Res Commun 480:499–507
Shan Q et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotech 31:686–688
Shi J et al (2016) ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. doi:10.1111/pbi.12603
Sternberg SH, Redding S, **ek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507:62–67
Sunilkumar G, Rathore KS (2001) Transgenic cotton: factors influencing Agrobacterium-mediated transformation and regeneration. Mol Breed 8:37–52
Sunilkumar G, Mohr L, Lopata-Finch E, Emani C, Rathore KS (2002) Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol Biol 50:463–479
Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS (2006) Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci 103:18054–18059
Tang W, Tang AY (2016) Applications and roles of the CRISPR system in genome editing of plants. J For Res. doi:10.1007/s11676-016-0281-7
Voytas DF, Gao C (2014) Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol 12:e1001877
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951
Wang W, Akhunova A, Chao S, Akhunov E (2016) Optimizing multiplex CRISPR/Cas9-based genome editing for wheat. bioRxiv. doi:10.1101/051342
Wong N, Liu W, Wang X (2015) WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol 16:1. doi:10.1186/s13059-015-0784-0
**e K, Zhang J, Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol Plant 7:923–926
**ng H-L et al (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327. doi:10.1186/s12870-014-0327-y
Xu R-F et al (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep 5:11491. doi:10.1038/srep11491
Yan R et al (2016) Progress in genome sequencing will accelerate molecular breeding in cotton (Gossypium spp.). 3 Biotech 6:217. doi:10.1007/s13205-016-0534-3
Zhang H et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhang T et al (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotech 33:531–537
Zhang Y et al (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:12617. doi:10.1038/ncomms12617
Acknowledgements
The base vectors for assembling the transformation constructs were kindly provided by Dr. Daniel Voytas, University of Minnesota.
Author contributions
KR: conception and design, acquisition of data, analysis and interpretation of data, drafting and revising the article. MJ: conception and design, performing the experiments, acquisition of data, analysis and interpretation of data, drafting and revising the article. LC: performing the experiments, acquisition of data.
Funding
This research was supported by funding from Texas A&M AgriLife Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Janga, M.R., Campbell, L.M. & Rathore, K.S. CRISPR/Cas9-mediated targeted mutagenesis in upland cotton (Gossypium hirsutum L.). Plant Mol Biol 94, 349–360 (2017). https://doi.org/10.1007/s11103-017-0599-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-017-0599-3