Abstract
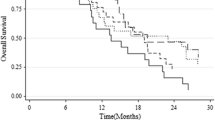
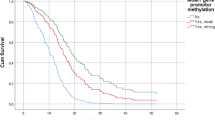
Resistance to temozolomide (TMZ) chemotherapy poses a significant challenge in the treatment of glioblastoma (GBM). Hypermethylation in O6-methylguanine-DNA methyltransferase (MGMT) promoter is thought to play a critical role in this resistance. Pyrosequencing (PSQ) has been shown to be accurate and robust for MGMT promoter methylation testing. The unresolved issue is the determination of a cut-off value for dichotomization of quantitative MGMT PSQ results into “MGMT methylated” and “MGMT unmethylated” patient subgroups as a basis for further treatment decisions. In this study, receiver operating characteristic (ROC) curve analysis was used to identify an optimal cutoff of MGMT promoter methylation by testing mean percentage of methylation of 4 CpG islands (76–79) within MGMT exon 1. The area under the ROC (AUC) as well as the best cutoff to classify the methylation were calculated. Positive likelihood ratio (LR+) was chosen as a diagnostic parameter for defining an optimal cut-off. Meanwhile, we also analyzed whether mean percentage of methylation at the investigated CpG islands could be regarded as a marker for evaluating prognostication. ROC analysis showed that the optimal threshold was 12.5% (sensitivity: 60.87%; specificity: 76%) in response to the largest LR+ 2.54. 12.5% was established to distinguish MGMT promoter methylation, which was confirmed using validation set. According to the cutoff value, the MGMT promoter methylation was found in 58.3% of GBM. Mean methylation level of the investigated CpG sites strong correlated with overall survival (OS), which means GBM patients with a high level of methylation survived longer than those with low level of methylation(log-rank test, P = 0.017). In conclusion, ROC curve analysis enables the best cutoff for discriminating MGMT promoter methylation status. LR+ can be used as a key factor that evaluates cutoff. The promoter methylation level of MGMT by PSQ in GBM patients had prognostic value.





Similar content being viewed by others
Abbreviations
- GBM:
-
Glioblastoma
- PSQ:
-
Pyrosequencing
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the ROC
- LR+:
-
Positive likelihood ratio
- OS:
-
Overall survival
- TMZ:
-
Temozolomide
- MGMT:
-
O6-methylguanine-DNA methyltransferase
- GTR:
-
Gross total resection
- PR:
-
Partial resection
- KPS:
-
Karnofsky Performance Scale
- HR:
-
Hazard ratio
- CI:
-
Confidence interval
References
Preusser M, de Ribaupierre S, Wöhrer A, Erridge SC, Hegi M, Weller M, Stupp R (2011) Current concepts and management of glioblastoma. Ann Neurol 70(1):9–21. doi:10.1002/ana.22425
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10):987–996. doi:10.1056/NEJMoa043330
Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR (2008) Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol 26(25):4189–4199. doi:10.1200/JCO.2007.11.5964
Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K (2011) A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol 121(5):651–661. doi:10.1007/s00401-011-0803-5
Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, Wong H, Liloglou T, Haylock B, Walker C (2009) Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer 101(1):124–131. doi:10.1038/sj.bjc.6605127
Uno M, Oba-Shinjo SM, Camargo AA, Moura RP, Aguiar PH, Cabrera HN, Begnami M, Rosemberg S, Teixeira MJ, Marie SK (2011) Correlation of MGMT promoter methylation status with gene and protein expression levels in glioblastoma. Clinics 66(10):1747–1755. doi:10.1590/S1807-59322011001000013
Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, Hegi ME (2010) MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 6(1):39–51. doi:10.1038/nrneurol.2009.197
Quillien V, Lavenu A, Karayan-Tapon L, Carpentier C, Labussière M, Lesimple T, Chinot O, Wager M, Honnorat J, Saikali S, Fina F, Sanson M, Figarella-Branger D (2012) Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer 118(17):4201–4211. doi:10.1002/cncr.27392
Hao X, Raymond T, Bin Y (2015) Detection of MGMT promoter methylation in glioblastoma using pyrosequencing. Int J Clin Exp Pathol 8(2):1790–1796
Sun Y, Li S, Shen K, Ye S, Cao D, Yang J (2015) DAPK1, MGMT and RARB promoter methylation as biomarkers for high-grade cervical lesions. Int J Clin Exp Pathol 8(11):14939–14945
Florkowski CM (2008) Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: communicating the performance of diagnostic tests. Clin Biochem Rev 29(Suppl1):83–87
Pernambuco L, Espelt A, Costa de Lima K (2016) Screening for Voice Disorders in Older Adults (RAVI)-Part III: Cutoff Score and Clinical Consistency. J Voice. doi:10.1016/j.jvoice.2016.03.003
Quillien V, Lavenu A, Sanson M, Legrain M, Dubus P, Karayan-Tapon L, Mosser J, Ichimura K, Figarella-Branger D (2014) Outcome-based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patients. J Neurooncol 116:487–496. doi:10.1007/s11060-013-1332-y
Preusser M, Berghoff AS, Manzl C, Filipits M, Weinhäusel A, Pulverer W, Dieckmann K, Widhalm G, Wöhrer A, Knosp E, Marosi C, Hainfellner JA (2014) Clinical Neuropathology practice news 1-2014: pyrosequencing meets clinical and analytical performance criteria for routine testing of MGMT promoter methylation status in glioblastoma. Clin Neuropathol 33(1):6–14. doi:10.5414/NP300730
Tateishi R, Yoshida H, Matsuyama Y, Mine N, Kondo Y, Omata M (2008) Diagnostic accuracy of tumor markers for hepatocellular carcinoma: a systematic review. Hepatol Int 2(1):17–30. doi:10.1007/s12072-007-9038-x
Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M, Combs SE, Vesper J, Braun C, Meixensberger J, Ketter R, Mayer-Steinacker R, Reifenberger G, Weller M, NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society (2012) Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol 13:707–715. doi:10.1016/S1470-2045(12)70164-X
Malmström A, Grønberg BH, Marosi C, Stupp R, Frappaz D, Schultz H, Abacioglu U, Tavelin B, Lhermitte B, Hegi ME, Rosell J, Henriksson R, Nordic Clinical Brain Tumour Study Group (NCBTSG) (2012) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol 13:916–926. doi:10.1016/S1470-2045(12)70265-6
Weller M, van den Bent M, Hopkins K, Tonn JC, Stupp R, Falini A, Cohen-Jonathan-Moyal E, Frappaz D, Henriksson R, Balana C, Chinot O, Ram Z, Reifenberger G, Soffetti R, Wick W, European Association for Neuro-Oncology (EANO) Task Force on Malignant Glioma (2014) EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol 15:e395–e403. doi:10.1016/S1470-2045(14)70011-7
Hsu CY, Ho HL, Lin SC, Chen MH, Hsu SP, Yen YS, Guo WY, Ho DM (2016) Comparative assessment of 4 methods to analyze MGMT status in a series of 121 glioblastoma patients. Appl Immunohistochem Mol Morphol. doi:10.1097/PAI.0000000000000331
Christians A, Hartmann C, Benner A, Meyer J, von Deimling A, Weller M, Wick W, Weiler M (2012) Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS ONE. doi:10.1371/journal.pone.0033449
Bienkowski M, Berghoff AS, Marosi C, Wöhrer A, Heinzl H, Hainfellner JA, Preusser M (2015) Practice guide 5-2015: MGMT methylation pyrosequencing in glioblastoma: unresolved issues and open questions. Clin Neuropathol 34(5):250–257. doi:10.5414/NP300904
Reifenberger G, Hentschel B, Felsberg J, Schackert G, Simon M, Schnell O, Westphal M, Wick W, Pietsch T, Loeffler M, Weller M, German Glioma Network (2012) Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer 131(6):1342–1350. doi:10.1002/ijc.-27385
Cabrini G, Fabbri E, Lo Nigro C, Dechecchi MC, Gambari R (2015) Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review). Int J Oncol 47(2):417–428. doi:10.3892/ijo.2015.3026
Hsu C-Y, Ho H-L, Lin S-C, Chang-Chien Y-C, Chen M-H, Hsu SP-C, Yen Y-S, Guo W-Y, Ho DM-T (2015) Prognosis of glioblastoma with faint MGMT methylation-specific PCR product. J Neurooncol 122:179–188. doi:10.1007/s11060-014-1701-1
Greiner M, Pfeiffer D, Smith RD (2000) Principles and practical application of the receiver operating characteristic analysis for diagnostic test. Prev Vet Med 45(1–2):23–41
Steurer J, Fischer JE, Bachmann LM, Koller M, ter Riet G (2002) Communicating accuracy of tests to general practitioners: a controlled study. BMJ 324(7341):824–826
Bandos AI, Rockette HE, Gur D (2010) Use of likelihood ratios for comparisons of binary diagnostic tests: underlying ROC curves. Med Phys 37(11):5821–5830
Biggerstaff BJ (2000) Comparing diagnostic tests: a simple graphic using likelihood ratios. Stat Med 19(5):649–663
Kim DC, Kim KU, Kim YZ (2016) Prognostic role of methylation status of the MGMT promoter determined quantitatively by pyrosequencing in glioblastoma patients. J Korean Neurosurg Soc 59(1):26–36. doi:10.3340/jkns.2016.59.1.26
Irwig L, Macaskill P, Glasziou P, Fahey M (1995) Meta-analytic methods for diagnostic test accuracy. J Clin Epidemiol 48:119–130 (discussion 31–2)
Swets JA (1988) Measuring the accuracy of diagnostic systems. Science 240:1285–1293
Begg CB (1991) Advances in statistical methodology for diagnostic medicine in the 1980’s. Stat Med 10:1887–1895
Brigliadori G, Foca F, Dall’Agata M, Rengucci C, Melegari E, Cerasoli S, Amadori D, Calistri D, Faedi M (2016) Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity in glioblastoma. J Neurooncol 128(2):333–339. doi:10.1007/s11060-016-2116-y
Collins VP, Ichimura K, Di Y, Pearson D, Chan R, Thompson LC, Gabe R, Brada M, Stenning SP (2014) Prognostic and predictive markers in recurrent high grade glioma; results from the BR12 randomised trial. Acta Neuropathol Commun. doi:10.1186/2051-5960-2-68
Acknowledgements
Funding was provided by Research Project of Chinese Society of Neuro-oncology (Grant No. CSNO-2013-MS008). The Project of Healty and Famliy Planing Commission of Gansu (Grant No. GSWSKY-2015-58/-2014-31), the Lanzhou Science and Technology Bureau Project (Grant No. 2013-3-27/2015-3-86), and the doctoral research fund of Lanzhou University Second Hospital (Grant No. ynbskyjj 2015-1-02/2015-2-11/2015-2-5)
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
We declare that we have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yuan, G., Niu, L., Zhang, Y. et al. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J Neurooncol 133, 193–201 (2017). https://doi.org/10.1007/s11060-017-2433-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-017-2433-9