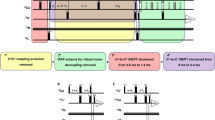
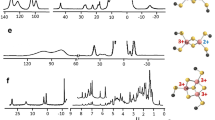
Abstract
Molecular size has limited solution NMR analyses of proteins. We report 13C–13C NOESY experiments on a 480 kDa protein, the multi-subunit ferritin nanocage with gated pores. By exploiting 13C-resonance-specific chemical shifts and spin diffusion effects, we identified 75% of the amino acids, with intraresidue C–C connectivities between nuclei separated by 1–4 bonds. These results show the potential of 13C–13C NOESY for solution studies of molecular assemblies >100 kDa.




Similar content being viewed by others
References
Banci L, Bertini I, Luchinat C (1991) Nuclear and electron relaxation. The magnetic nucleus-unpaired electron coupling in solution. VCH, Weinheim
Bermel W, Bertini I, Duma L, Emsley L, Felli IC, Pierattelli R, Vasos PR (2005) Complete assignment of heteronuclear protein resonances by protonless NMR spectroscopy. Angew Chem Int Ed 44:3089–3092
Bermel W, Bertini I, Felli IC, Kümmerle R, Pierattelli R (2003) 13C direct detection experiments on the paramagnetic oxidized monomeric copper, zinc superoxide dismutase. J Am Chem Soc 125:16423–16429
Bermel W, Bertini I, Felli IC, Lee Y-M, Luchinat C, Pierattelli R (2006a) Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J Am Chem Soc 128:3918–3919
Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R (2006b) 13C-detected protonless NMR spectroscopy of proteins in solution. Progr NMR Spectrosc 48:25–45
Bertini I, Duma L, Felli IC, Fey M, Luchinat C, Pierattelli R, Vasos PR (2004a) A heteronuclear direct detection NMR experiment for protein backbone assignment. Angew Chem Int Ed 43:2257–2259
Bertini I, Felli IC, Kümmerle R, Luchinat C, Pierattelli R (2004b) 13C–13C NOESY: a constructive use of 13C–13C spin-diffusion. J Biomol NMR 30:245–251
Bertini I, Felli IC, Kümmerle R, Moskau D, Pierattelli R (2004c) 13C–13 C NOESY: an attractive alternative to study large macromolecules. J Am Chem Soc 126:464–465
Bertini I, Fragai M, Luchinat C, Parigi G (2000) 1H NMRD profiles of diamagnetic proteins: a model-free analysis. Magn Reson Chem 38:543–550
Brüschweiler R and Ernst RR (1992) Molecular dynamics monitored by cross-correlated cross relaxation of spins quantized along orthogonal axes. J Chem Phys 96:1758–1766
Dalvit C (1992) 1H to 15N polarization transfer via 1H chemical-shift anisotropy-1H–15N dipole dipole cross corelation. J Magn Reson 97:645–650
Fernández C, Wider G (2003) TROSY in NMR studies of the structure and function of large biological macromolecules. Curr Opin Struct Biol 13:570–580
Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K (2004) Uniform and residue-specific 15N-labeling of proteins on a highly deuterated background. J Biomol NMR 29:289–297
Fischer MWF, Zeng L, Zuiderweg ERP (1996) Use of 13C–13C NOE for the assignment of NMR lines of larger labeled proteins at larger magnetic fields. J Am Chem Soc 118:12457–12458
Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K (2002) NMR analysis of a 900 KDa GroEL GROES complex. Nature 418:207–211
Frueh DP, Sun ZYJ, Vosburg DA, Walsh CT, Hoch JC, Wagner G (2006) Non-uniformly sampled double-TROSY hNcaNH experiments for NMR sequential assignments of large proteins. J Am Chem Soc 128:5757–5763
Ha Y, Shi DS, Small GW, Theil EC, Allewell NM (1999) Crystal structure of bullfrog M ferritin at 2.8 A resolution: analysis of subunit interactions and the binuclear metal center. J Biol Inorg Chem 4:243–256
Horst R, Wider G, Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K (2006) Proton–proton Overhauser NMR spectroscopy with polypeptide chains in large structures. Proc Natl Acad Sci USA 103:15445–15450
Kaptein R, Zuiderweg ERP, Scheek RM, Boelens R, van Gunsteren WF (1985) A protein structure from nuclear magnetic resonance data: lac Repressor headpiece. J Mol Biol 182:179–182
Liu X, Theil EC (2005) Ferritins: dynamic management of biological iron and oxygen chemistry. Acc Chem Res 38:167–175
Miclet E, Williams Jr DC, Clore GM, Bryce DL, Boisbouvier J, Bax A (2004) Relaxation-optimized NMR spectroscopy of methylene groups in proteins and nucleic acids. J Am Chem Soc 126: 10560–10570
Neuhaus D, Williamson M (1989) The nuclear Overhauser effect in structural and conformational analysis. VCH, New York
Pervushin K, Riek R, Wider G, Wüthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Riek R, Wider G, Pervushin K, Wüthrich K (1999) Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc Natl Acad Sci USA 96:4918–4923
Trikha J, Theil EC, Allewell NM (1995) High resolution crystal structures of amphibian red-cell L ferritin: potential roles for structural plasticity and solvation in function. J Mol Biol 248:949–967
Tugarinov V, Choy WY, Orekhov VY, Kay LE (2005a) Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc Natl Acad Sci USA 102: 622–627
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced 1H–13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428
Tugarinov V, Ollerenshaw JE, Kay LE (2005b) Probing side-chain dynamics in high molecular weight proteins by deuterium NMR spin relaxation. An application to a 82-kDa enzyme. J Am Chem Soc 127:8214–8225
Wider G (2005) NMR techniques used with very large biological macromolecules in solution. Methods Enzymol 394:382–398
Williamson MP, Havel TF, Wüthrich K (1985) Solution Conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol 182:295–315
Acknowledgements
The work was supported in part by MIUR (COFIN 2005), NIH grant DK20251 (ECT and MM) and the CHRCO Foundation (ECT). The authors are grateful to Dr. **aofeng Liu for advice on recombinant ferritin structure/function and expression and to Dr. Rainer Kümmerle for the competent insights on spectral acquisition.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Matzapetakis, M., Turano, P., Theil, E.C. et al. 13C–13C NOESY spectra of a 480 kDa protein: solution NMR of ferritin. J Biomol NMR 38, 237–242 (2007). https://doi.org/10.1007/s10858-007-9163-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-007-9163-9