Abstract
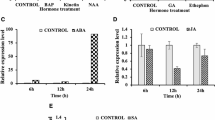
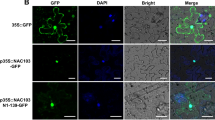
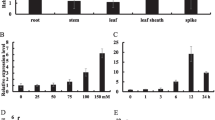
OsNAC45 belongs to the NAC family of (for NAM/ATAF1/2/CUC2) transcription factors, which play an important role in plant development and abiotic stress responses. Transgenic rice plants overexpressing OsNAC45 have exhibited improved tolerance to salt and drought stress. This study aimed to investigate the mechanism underlying the effects of OsNAC45 on root development under abiotic stress in rice Zhonghua11 (ZH11) (Oryza sativa L.). The OsNAC45 GUS expression was strong in roots of the adult stage, weak in the seedling stage, and induced by low-temperature stress in the seedling stage. Under NaCl and low temperature stress, the OsNAC45 overexpression (OE) lines exhibited maintenance of root structure and normal growth of the seedling root, but the RNAi lines of OsNAC45 were more sensitive. The roots of the OE lines accumulated less superoxide anion under low temperature stress and more hydrogen peroxide under normal conditions than WT. After NaCl treatment, the Peroxidase (POD) assay showed POD activity of the roots in OE lines were elevated, but were decreased in RNAi lines and WT; additionally, the increment of lignin content in OE lines was significantly higher than RNAi lines and WT in roots. Furthermore, expression levels of some rice root development-related genes were altered in the transgenic plants. Under cold and salt stresses, expression of NCED4, a key enzyme in abscisic acid synthesis, was decreased in OE lines and raised in RNAi lines compared to WT. The data suggests that OsNAC45 may participate in the response of cold/salt stresses by lessening the accumulation of reactive oxygen species and increasing lignin in rice roots thus alleviating the restraint to the growth of seedling root. Overexpression of OsNAC45 weakened the high temperature tolerance; however, inhibiting its expression could improve the tolerance. Together, these results indicate that OsNAC45 plays an important role in the coordination, development, and response of rice plants under abiotic stresses.







Similar content being viewed by others
References
An X, Liao Y, Zhang J, Dai L, Zhang N, Wang B et al (2015) Overexpression of rice NAC gene SNAC1 in ramie improves drought and salt tolerance. Plant Growth Regul 76(2):211–223
Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B et al (2010) SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22(3):640–654
Fang Y, You J, **e K, **e W, **ong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genom 280:547–563
Fang Y, Liao K, Du H, Xu Y, Song H, Li X et al (2015) A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot 66(21):6803–6817
Fryer MJ, Oxborough K, Mullineaux PM, Baker NR (2002) Imaging of photo-oxidative stress responses in leaves. J Exp Bot 53(372):1249–1254
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M et al (2004) A dehydration induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Furuta KM, Yadav SR, Lehesranta S, Belevich I, Miyashima S, Heo JO et al (2014) Plant development: Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science 345(6199):933–937
Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH et al (2004) Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol 135:773–782
Guo J, Wang F, Song J, Sun W, Zhang XS (2010) The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231(2):293–303
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways,is involved in salt stress response and lateral root development. Plant J 44(6):903–916
Hu H, Dai M, Yao J, **ao B, Li X, Zhang Q,et al (2006) Overexpressing a NAM, ATAF and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y,et al (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW et al (2013) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotech J 11:101–114
Jia L, Zhang B, Mao C, Li J, Wu Y, Wu P et al (2008) OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativaL.). Planta 228:51–59
Kadier Y, Zu Y, Dai Q, Song G, Lin S, Sun Q et al (2017) Genome-wide identification, classification and expression analysis of NAC family of genes in sorghum [Sorghum bicolor (L.) Moench]. Plant Growth Regul 83(2):301–312
Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH et al. (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323:1053–1057
Lee Y, Rubio MC, Alassimone J, Geldner N (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153(2):402–412
Lin C, Kao C (2001) Cell wall peroxidase activity, hydrogen peroxide level and NaCl-inhibited root growth of rice seedlings. Plant Soil 230:135–143
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 22DDCT method. Methods 25:402–408
Ma B, Yin CC, He SJ, Lu X, Zhang WK, Lu TG, Chen SY, Zhang JS (2014) Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet 10(10):e1004701
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17(11):2993–3006
Moura JC, Bonine CA, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52(4):360–376
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819(2):97–103
Niu F, Wang C, Yan J, Guo X, Wu F, Yang B,et al (2016) Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napusL.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol Biol 92(1–2):89–104
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H,et al (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465(1–2):30–44
Nuruzzaman M, Sharoni A, Satoh K, Karim MR, Harikrishna JA, Shimizu T et al (2015) NAC transcription factor family genes are differentially expressed in rice during infections with Rice dwarf virus, Rice black-streaked dwarf virus, Rice grassy stunt virus, Rice ragged stunt virus and Rice transitory yellowing virus. Front Plant Sci 6:676. https://doi.org/10.3389/fpls.2015.00676
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10(2):79–87
Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17(6):369–381
Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19(7):2197–2212
Sakuraba Y, Kim YS, Han SH, Lee BD, Paek NC (2015a) The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27(6):1771–1787
Sakuraba Y, Piao W, Lim JH, Han SH, Kim YS, An G et al (2015b) Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol 56(12):2325–2339
Sengupta D, Naik D, Reddy AR (2015) Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: a structure-function update. J Plant Physiol 179:40–55
Shen H, Yin Y, Chen F, Xu Y, Dixon RA (2009) Bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. BioEnergy Res 2:217–232
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85(2):159–170
Wang HZ, Dixon RA (2012) On-off switches for secondary cell wall biosynthesis. Mol Plant 5(2):297–303
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B et al (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290
**e Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
**ong Y, Liu T, Tian C, Sun S, Li J, Chen M (2005) Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol 59:191–203
Xu ZY, Kim SY, Hyeon do Y, Kim DH, Dong T, Park Y et al (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25(11):4708–4824
Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K (1992) Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol 33:217–224
Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23:2155–2168
Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL (2014) The Membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet 10(3):e1004243
You J, Chan Z (2015) ROS regulation during abiotic stress responses in crop plants. Front Plant Sci 6:1092. https://doi.org/10.3389/fpls.2015.01092
Yu X, Liu Y, Wang S, Tao Y, Wang Z, Mijiti A et al (2016) A chickpea stress-responsive NAC transcription factor, CarNAC5, confers enhanced tolerance to drought stress in transgenic Arabidopsis. Plant Growth Regul 79(2):187–197
Zhao L, Hu Y, Chong K, Wang T (2010) ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Ann Bot 105(3):401–409
Zheng X, Chen B, Lu G, Han B (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379:985–989
Zhong R, Lee C, Ye ZH (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3:1087–1103
Zhou Y, Huang W, Liu L, Chen T, Zhou F, Lin Y (2013) Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol 13:132. https://doi.org/10.1186/1471-2229-13-132
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (30900070), National Key Technologies R&D Program of China (2016ZX08001003-004) and Shanghai Youth Science Talents to Sail Plan (15YF1410600).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, S., Huang, A., Li, J. et al. OsNAC45 plays complex roles by mediating POD activity and the expression of development-related genes under various abiotic stresses in rice root. Plant Growth Regul 84, 519–531 (2018). https://doi.org/10.1007/s10725-017-0358-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-017-0358-0