Abstract
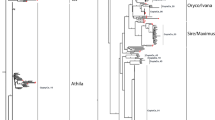
Peanut is an allotetraploid (2n = 2x = 40, AABB) of recent origin. Arachis duranensis and A. ipaënsis, the most probable diploid ancestors of the cultigen, and several other wild diploid species with different genomes (A, B, D, F and K) are used in peanut breeding programs. However, the genomic relationships and the evolutionary pathways of genome differentiation of these species are poorly understood. We performed a sequence-based phylogenetic analysis of the L1 reverse transcriptase and estimated its representation and chromosome distribution in species of five genomes and three karyotype groups with the aim of contributing to the knowledge of the genomic structure and evolution of peanut and wild diploid relatives. All the isolated rt fragments were found to belong to plant L1 lineage and were named ALI. The best supported phylogenetic groups were not concordant with the genomes or karyotype groups. The copy number of ALI sequences was higher than the expected one for plants and directly related to genome size. FISH experiments revealed that ALI is mainly located on the euchromatin of interstitial and distal regions of most chromosome arms. Divergence of ALI sequences would have occurred before the differentiation of the genomes and karyotype groups of Arachis. The representation and chromosome distribution of ALI in peanut was almost additive of those of the parental species suggesting that the spontaneous hybridization of the two parental species of peanut followed by chromosome doubling would not have induced a significant burst of ALI transposition.







Similar content being viewed by others
Abbreviations
- LINEs:
-
Long interspersed nuclear elements
- rt :
-
Reverse transcriptase
- GSS:
-
Genomic survey sequences
- ESTs:
-
Expressed sequence tags
- FISH:
-
Fluorescent in situ hybridization
- LTR:
-
Long terminal repeats
- SINEs:
-
Short interspersed nuclear elements
- NJ:
-
Neighbor joining
- GISH:
-
Genome in situ hybridization
- ORFs:
-
Open reading frames
- PCR:
-
Polymerase chain reaction
- dNTPs:
-
Deoxynucleotide triphosphates
- CSPD:
-
Disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro)tricyclo [3.3.1.13,7]decan}-4-yl)phenyl phosphate
- DAPI:
-
4,6-Diamidino-2-phenylindole
- TE:
-
Transposable element
References
Alix K, Heslop-Harrison JS (2004) The diversity of retroelements in diploid and allotetraploid Brassica species. Plant Mol Biol 54:895–909
Altschul SF, Gish W, Miller W, Myers E, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Bertioli DJ, Moretzsohn MC, Madsen LH, Sandal N, Leal-Bertioli SC, Guimarães PM, Hougaard BK, Fredslund J, Schauser L, Nielsen AM, Sato S, Tabata S, Cannon SB, Stougaard J (2009) An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genome 10:45
Bertioli DJ, Vidigal B, Nielen S, Ratnaparkhe MB, Lee T-H, Leal-Bertioli SCM, Kim C, Guimarães PM, Seijo G, Schwarzacher T, Paterson AH, Heslop-Harrison P, Araujo ACG (2013) The repetitive component of the A genome of peanut (Arachis hypogaea) and its role in remodelling intergenic sequence space since its evolutionary divergence from the B genome. Ann Bot 112:545–559
Burow MD, Simpson CE, Starr JL, Paterson A (2001) Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): broadening the gene pool of a monophyletic polyploid species. Genetics 159:823–837
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Di Rienzo JA, Casanoves F, Balzarini MG, González L, Tablada M, Robledo CW (2013) InfoStat version 2014. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. http://www.infostat.com.ar/
Doležel J, Bartos J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot 95:99–110
Doolittle RF, Feng DF, Johnson MS, McClure MA (1989) Origins and evolutionary relationships of retroviruses. Quart Rev Biol 64:1–30
Fernández A, Krapovickas A (1994) Cromosomas y evolución en Arachis (Leguminosae). Bonplandia 8:188–220
Flavell AJ (1995) Retroelements, reverse transcriptase and evolution. Comp Biochem Physiol B Biochem Mol Biol 110:3–15
Grabiele M, Chalup L, Robledo G, Seijo G (2012) Genetic and geographic origin of domesticated peanut as evidenced by 5S rDNA and chloroplast DNA sequences. Plant Syst Evol 298:1151–1165
Grattapaglia D, Sederoff R (1994) Genetic linkage Maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: map** strategy and RAPD markers. Genetics 137:1121–1137
Gregory MP, Gregory WC (1979) Exotic germoplasm of Arachis L. interspecific hybrids. J Hered 70:185–193
Hammons RO (1994) The origin and history of the groundnut. In: Smartt J (ed) The groundnut crop: a scientific basis for improvement. Chapman & Hall, London, pp 24–39
Han JS (2010) Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions. Mobile DNA 1:15
Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16:1252–1261
Heitkam T, Schmidt T (2009) BNR-a LINE family from Beta vulgaris-contains a RRM domain in open reading frame 1 and defines a L1 sub-clade present in diverse plant genomes. Plant J 59:872–882
Husted L (1936) Cytological studies on the peanut Arachis. II. Chromosome number, morphology and behavior, and their application to the problem of the origin of the cultivated forms. Cytologia 7:396–423
Ichimura S, Mita K, Sugaya K (1997) A major non-LTR retrotransposon of Bombyx mori, L1Bm. J Mol Evol 45:253–264
Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160:1651–1659
Kochert G, Stalker HT, Gimenes M, Galgaro L, Lopes CR, Moore K (1996) RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae). Am J Bot 83:1282–1291
Krapovickas A, Gregory W (1994) Taxonomía del género Arachis (Leguminosae). Bonplandia 8:1–186
Kubis SE, Heslop-Harrison JS, Desel C, Schmidt T (1998) The genomic organization of non-LTR retrotransposons (LINEs) from three Beta species and five other angiosperms. Plant Mol Biol 36:821–831
Kubo Y, Okazaki S, Anzai T, Fujiwara H (2001) Structural and phylogenetic analysis of TRAS, telomeric repeat-specific non-LTR retrotransposon families in Lepidopteran insects. Mol Biol Evol 18:848–857
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Leeton PR, Smyth DR (1993) An abundant LINE-like element amplified in the genome of Lilium speciosum. Mol Gen Genet 237:97–104
Lim KY, Kovarik A, Matyasek R, Chase MW, Clarkson JJ, Grandbastien MA, Leitch AR (2007) Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol 175:756–763
Liu B, Wendel JF (2000) Retroelement activation followed by rapid repression in interspecific hybrid plants. Genome 43:874–880
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge RW, Martienssen R, Comai L (2005) Genomic changes in synthetic Arabidopsis polyploids. Plant J 41:221–230
Malik HS, Burke WD, Eickbush TH (1999) The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 16:793–805
Martin SL, Bushman FD (2001) Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol 2:467–475
Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11:1660–1676
Moretzhon MC, Gouvea EG, Inglis PW, Leal-Bertioli SCM, Valls JFM, Bertioli DJ (2013) A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Ann Bot 111:113–126
Moretzsohn MC, Barbosa AV, Alves-Freitas DM, Teixeira C, Leal-Bertioli SC, Guimarães PM, Pereira RW, Lopes CR, Cavallari MM, Valls JF, Bertioli DJ, Gimenes MA (2009) A linkage map for the B-genome of Arachis (Fabaceae) and its synteny to the A-genome. BMC Plant Biol 9:40
Moscone E, Matzke M, Matzke A (1996) The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105:231–236
Nielen S, Campos-Fonseca F, Leal-Bertioli S, Guimaraes P, Seijo G, Town C, Arrial R, Bertioli D (2009) FIDEL—a retrovirus-like retrotransposon and its distinct evolutionary histories in the A- and B-genome components of cultivated peanut. Chromosome Res 18:227–246
Nielen S, Vidigal B, Leal-Bertioli S, Ratnaparkhe M, Paterson A, Garsmeur O, D’Hont A, Guimarães P, Bertioli D (2011) Matita, a new retroelement from peanut: characterization and evolutionary context in the light of the Arachis A–B genome divergence. Mol Genet Genomics 287:21–38
Noma K, Ohtsubo E, Ohtsubo H (1999) Non-LTR retrotransposons (LINEs) as ubiquitous components of plant genomes. Mol Gen Genet 261:71–79
Ohshima K, Hamada M, Terai Y, Okada N (1996) The 3′-ends of tRNA-derived short interspersed repetitive elements are derived from the 3′-ends of long interspersed repetitive elements. Mol Cell Biol 16:3756–3764
Petrov DA (2002) Mutational equilibrium model of genome size evolution. Theor Popul Biol 61:531–543
Robledo G, Seijo JG (2008) Characterization of Arachis D genome using physical map** of heterochromatic regions and rDNA loci by FISH. Genet Mol Biol 31:717–724
Robledo G, Seijo G (2010) Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH map** of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theor Appl Genet 121:1033–1046
Robledo G, Lavia GI, Seijo G (2009) Species relations among wild Arachis species with the A genome as revealed by FISH map** of rDNA loci and heterochromatin detection. Theor Appl Genet 118:1295–1307
Rossi M, Gonçalves Araujo P, Van Sluys MA (2001) Survey of transposable elements in sugarcane expressed sequence tags (ESTs). Genet Mol Biol 24:147–154
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakamoto K, Ohmido N, Fukui K, Kamada H, Satoh S (2000) Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol Biol 44:723–732
Schmidt T (1999) LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes. Plant Mol Biol 40:903–910
Schön I, Arkhipova IR (2006) Two families of non-LTR retrotransposons, Syrinx and Daphne, from the Darwinulid ostracod, Darwinula stevensoni. Gene 371:296–307
Schwarzacher T, Ambros P, Schweizer D (1980) Application of Giemsa banding to orchid karyotype analysis. Plant Syst Evol 134:293–297
Schwarz-Sommer Z, Leclercq L, Göbel E, Saedler H (1987) Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J 6:3873–3880
Seijo G, Lavia GI, Fernández A, Krapovickas A, Ducasse D, Moscone EA (2004) Physical map** of 5S and 18S-25S rRNA genes evidences that Arachis duranensis and A. ipaënsis are the wild diploid species involved in the origin of A. hypogaea (Leguminosae). Am J Bot 91:2293–2303
Seijo G, Lavia GI, Fernández A, Krapovickas A, Ducasse D, Bertioli DJ, Moscone EA (2007) Genomic relationships between the cultivated peanut (Arachis hypogaea-Leguminosae) and its close relatives revealed by double GISH. Am J Bot 94:1963–1971
Shirasawa K, Bertioli D, Varshney R, Moretzsohn M, Leal-Bertioli S, Thudi M, Pandey M, Rami J, Foncéka D, Gowda M, Qin H, Guo B, Hong Y, Liang X, Hirakawa H, Tabata S, Isobe S (2013) Integrated consensus map of cultivated peanut and wild relatives reveals structures of the A and B genomes of Arachis and divergence of the legume genomes. DNA Res 20:173–184
Silvestri MC, Ortiz AM, Lavia GI (2014) rDNA loci and heterochromatin positions support a distinct genome type for ‘x = 9 species’ of section Arachis (Arachis, Leguminosae). Plant Syst Evol. doi:10.1007/s00606-014-1092-y
Simpson CE, Krapovickas A, Valls JFM (2001) History of Arachis including evidence of A. hypogaea L. progenitors. Peanut Sci 28:78–80
Smartt J, Gregory WC, Gregory MP (1978) The genomes of Arachis hypogaea. 1. Cytogenetic studies of putative genome donors. Euphytica 27:665–675
Smyshlyaev G, Voigt F, Blinov A, Barabas O, Novikova O (2013) Acquisition of an Archaea-like ribonuclease H domain by plant L1 retrotransposons supports modular evolution. Proc Natl Acad Sci USA 110:20140–20145
Stalker HT (1991) A new species-section Arachis of peanuts with D genome. Am J Bot 78:630–637
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vaio M, Mazzella C, Porro V, Speranza P, López-Carro B, Estramil E, Folle GA (2007) Nuclear DNA content in allopolyploid species and synthetic hybrids in the grass genus Paspalum. Plant Syst Evol 265:109–121
Valls JFM, Simpson CE (2005) New species of Arachis (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia 14:35–63
Vershinin AV, Druka A, Alkhimova AG, Kleinhofs A, Heslop-Harrison JS (2002) LINEs and gypsy-like retrotransposons in Hordeum species. Plant Mol Biol 49:1–14
Wenke T, Döbel T, Sörensen TR, Junghans H, Weisshaar B, Schmidt T (2011) Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell 23:3117–3128
Wessler SR (2006) Transposable elements and the evolution of eukaryotic genomes. Proc Natl Acad Sci USA 103:17600–17601
Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu ZD, Dubcovsky J, Keller B (2003) Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15:1186–1197
**ong Y, Eickbush TH (1988) Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol 5:675–690
**ong Y, Eickbush TH (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362
Zupunski V, Gubensek F, Kordis D (2001) Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol Biol Evol 18:1849–1863
Acknowledgments
This work is part of S. Samoluk Doctoral Thesis that will be presented in the Facultad de Ciencias Agrarias, Universidad Nacional de Rosario (Santa Fe, Argentina). It was supported by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Argentina, Project PICT 2007-1356 and PICT 2012-1875, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina, Project PIP No 11220090100613 and Universidad Nacional del Nordeste, PI No 189. S. Samoluk, M. Podio and L. Chalup received fellowships from CONICET. G. Robledo, S. Pessino, J.P.A. Ortiz and J. G. Seijo are research staff members of CONICET. We would like to thank INTA Manfredi Station, Córdoba, Argentina and the Texas Agriculture Experimental Station, Stephenville, Texas, for providing seeds of some accessions.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2015_9820_MOESM1_ESM.pdf
Alignments of deduced amino acid rt sequences used for construction of NJ trees of ALI elements and LINEs from other species. Arabidopsis thaliana Ta11-1 (L47193), Beta vulgaris BvL2 (FM993987), Zea mays Cin4 (Y00086), Hordeum vulgare BLIN (AJ270056), Cannabis sativa LINE-CS (AB013908), Rattus norvegicus L1 rat (U83119), Homo sapiens L1 hs (U93574), Drosophila melanogaster I (M14954), Drosophila melanogaster Jockey (M22874), Gallus gallus CR1 (U88211). The symbols “-, ? and *” indicates gaps, incomplete codons and stop codons, respectively. (PDF 140 kb)
Rights and permissions
About this article
Cite this article
Samoluk, S.S., Robledo, G., Podio, M. et al. First insight into divergence, representation and chromosome distribution of reverse transcriptase fragments from L1 retrotransposons in peanut and wild relative species. Genetica 143, 113–125 (2015). https://doi.org/10.1007/s10709-015-9820-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-015-9820-y