Abstract
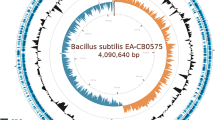
Bacillus subtilis XF-1 is a gram-positive, plant-associated bacterium that stimulates plant growth and produces secondary metabolites that suppress soil-borne plant pathogens. In particular, it is especially highly efficient at controlling the clubroot disease of cruciferous crops. Its 4,061,186-bp genome contains an estimated 3853 protein-coding sequences and the 1155 genes of XF-1 are present in most genome-sequenced Bacillus strains: 3757 genes in B. subtilis 168, and 1164 in B. amyloliquefaciens FZB42. Analysis using the Cluster of Orthologous Groups database of proteins shows that 60 genes control bacterial mobility, 221 genes are related to cell wall and membrane biosynthesis, and more than 112 are genes associated with secondary metabolites. In addition, the genes contributed to the strain’s plant colonization, bio-control and stimulation of plant growth. Sequencing of the genome is a fundamental step for develo** a desired strain to serve as an efficient biological control agent and plant growth stimulator. Similar to other members of the taxon, XF-1 has a genome that contains giant gene clusters for the non-ribosomal synthesis of antifungal lipopeptides (surfactin and fengycin), the polyketides (macrolactin and bacillaene), the siderophore bacillibactin, and the dipeptide bacilysin. There are two synthesis pathways for volatile growth-promoting compounds. The expression of biosynthesized antibiotic peptides in XF-1 was revealed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry.




Similar content being viewed by others
References
Asaka O, Shoda M (1996) Biocontrol of Rhizoctonia solani dam**-off of tomato with Bacillus subtilis RB14. Appl Environ Microbiol 62(11):4081–4085
Atkins T, Prior R, Mack K, Russell P, Nelson M, Oyston P, Dougan G, Titball R (2002) A mutant of Burkholderia pseudomallei, auxotrophic in the branched chain amino acid biosynthetic pathway, is attenuated and protective in a murine model of melioidosis. Infect Immun 70(9):5290
Badger JH, Olsen GJ (1999) CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol 16(4):512
Bais HP, Fall R, Vivanco JM (2004) Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134(1):307–319
Bird C, Wyman M (2003) Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression. Appl Environ Microbiol 69(12):7009–7018
Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R (2004) Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol 186(12):3970–3979
Buczacki S, Moxham SE (1983) Structure of the resting spore wall of Plasmodiophora brassicae revealed by electron microscopy and chemical digestion. T Brit Mycol Soc 81(2):221–231
Candela T, Fouet A (2006) Poly-gamma-glutamate in bacteria. Mol Microbiol 60(5):1091–1098
Cassán F, Maiale S, Masciarelli O, Vidal A, Luna V, Ruiz O (2009) Cadaverine production by Azospirillum brasilense and its possible role in plant growth promotion and osmotic stress mitigation. Eur J Soil Biol 45(1):12–19
Cavaglieri L, Orlando J, Rodriguez M, Chulze S, Etcheverry M (2005) Biocontrol of Bacillus subtilis against Fusarium verticillioides in vitro and at the maize root level. Res Microbiol 156(5):748–754
Chen L, Helmann JD (1994) The Bacillus subtilis sigma D-dependent operon encoding the flagellar proteins FliD, FliS, and FliT. J Bacteriol 176(11):3093–3101
Chen XH, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter AW (2006) Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol 188(11):4024–4036
Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R (2009) Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol 140(1–2):27–37
Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, Morgenstern B, Voss B, Hess WR, Reva O, Junge H, Voigt B, Jungblut PR, Vater J, Süssmuth R, Liesegang H, Strittmatter A, Gottschalk G, Borriss R (2007) Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol 25(9):1007–1014
Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71(9):4951–4959
Cox GN, Kusch M, Edgar RS (1981) Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J Cell Biol 90(1):7–17
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23(6):673
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acid Res 27(23):4636
Duckworth OW, Bargar JR, Sposito G (2009) Coupled biogeochemical cycling of iron and manganese as mediated by microbial siderophores. Biometals 22(4):605–613
Duffy BK, Défago G (1999) Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 65(6):2429–2438
Fan B, Carvalhais LC, Becker A, Fedoseyenko D, von Wirén N, Borriss R (2012) Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol 12(1):116
Ferreira J, Matthee F, Thomas A (1991) Biological control of Eutypa lata on grapevine by an antagonistic strain of Bacillus subtilis. Phytopathology 81(3):283–287
Fredrick KL, Helmann JD (1994) Dual chemotaxis signaling pathways in Bacillus subtilis: a sigma D-dependent gene encodes a novel protein with both CheW and CheY homologous domains. J Bacteriol 176(9):2727–2735
Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60(4):579–598
He P, Hao K, Blom J, Rückert C, Vater J, Mao Z, Wu Y, Hou M, He P, He Y (2013) Genome sequence of the plant growth promoting strain Bacillus amyloliquefaciens subsp. plantarum B9601-Y2 and expression of mersacidin and other secondary metabolites. J Biotechnol 164(2):281–291
Hecker M, Völker U (2004) Towards a comprehensive understanding of Bacillus subtilis cell physiology by physiological proteomics. Proteomics 4(12):3727–3750
Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF (2002) The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun 70(9):5246–5255
Holtmann G, Bakker EP, Uozumi N, Bremer E (2003) KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol 185(4):1289–1298
Huber DM (1980) The role of mineral nutrition in defense. In: Horsfall JG, Cowling EB (eds) Plant Disease. AcademicPress, New York, pp 381–406
Jacques P (2011) Surfactin and other lipopeptides from Bacillus spp. In: Soberón-Chávez G (ed) Biosurfactants. Springer, Berlin, pp 57–91
Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T (2008) KEGG for linking genomes to life and the environment. Nucleic Acid Res 36(suppl 1):480–484
Kearns DB, Chu F, Branda SS, Kolter R, Losick R (2004) A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55(3):739–749
Kearns DB, Chu F, Rudner R, Losick R (2004) Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52(2):357–369
Kent WJ (2002) BLAT-the BLAST-like alignment tool. Genome Res 12(4):656
Kilian M, Steiner U, Krebs B, Junge H, Schmiedeknecht G, Hain R (2000) FZB24® Bacillus subtilis–mode of action of a microbial agent enhancing plant vitality. Pflanzenschutz-Nachrichten Bayer 1(1):72–93
Kirby JR, Kristich CJ, Saulmon MM, Zimmer MA, Garrity LF, Zhulin IB, Ordal GW (2001) CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol Microbiol 42(3):573–585
Kloepper JW, Leong J, Teintze M, Schroth MN (1980) Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286(5776):885–886
Kloepper JW, Ryu C-M, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94(11):1259–1266
Kobayashi K (2007) Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol 66(2):395–409
Koumoutsi A, Chen X-H, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186(4):1084–1096
Lazarevic V, Soldo B, Médico N, Pooley H, Bron S, Karamata D (2005) Bacillus subtilis α-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol 71(1):39–45
Leclère V, Béchet M, Adam A, Guez JS, Wathelet B, Ongena M, Thonart P, Gancel F, Chollet-Imbert M, Jacques P (2005) Mycosubtilin overproduction by Bacillus subtilis BBG100 enhances the organism’s antagonistic and biocontrol activities. Appl Environ Microbiol 71(8):4577–4584
Liu Q, **ong G, Mao Z, Wu Y, He Y (2012) Analyses for the colonization ability of Bacillus subtilis XF-1 in the rhizosphere. Acta phytophylacica sinica 39:425–430
Lopez D, Kolter R (2010) Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev 34(2):134–149
Lopez D, Vlamakis H, Kolter R (2009) Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev 33(1):152–163
Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P (2001) Early steps of Bacillus subtilis primosome assembly. J Biol Chem 276(49):45818
Marvasi M, Visscher PT, Casillas Martinez L (2010) Exopolymeric substances (EPS) from Bacillus subtilis : polymers and genes encoding their synthesis. FEMS Microbiol Lett 313(1):1–9
Merrick M, Edwards R (1995) Nitrogen control in bacteria. Microbiol Rev 59(4):604–622
Mirel DB, Lauer P, Chamberlin MJ (1994) Identification of flagellar synthesis regulatory and structural genes in a sigma D-dependent operon of Bacillus subtilis. J Bacteriol 176(15):4492–4500
Moxham SE, Buczacki S (1983) Chemical composition of the resting spore wall of Plasmodiophora brassicae. T British Mycol Soc 80(2):297–304
Neilands J (1995) Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270(45):26723–26726
Noguchi H, Park J, Takagi T (2006) MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acid Res 34(19):5623
Omoike A, Chorover J (2004) Spectroscopic study of extracellular polymeric substances from Bacillus subtilis: aqueous chemistry and adsorption effects. Biomacromolecules 5(4):1219–1230
** L, Boland W (2004) Signals from the underground: bacterial volatiles promote growth in Arabidopsis. Trends Plant Sci 9(6):263–266
Rao CV, Glekas GD, Ordal GW (2008) The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol 16(10):480–487
Ryu CM, Farag MA, Hu C-H, Reddy MS, Wei HX, Paré PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. P Natl A Sci 100(8):4927–4932
Schultz D, Wolynes PG, Jacob EB, Onuchic JN (2009) Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. P Natl A Sci 106(50):21027
Sekowska A, Bertin P, Danchin A (1998) Characterization of polyamine synthesis pathway in Bacillus subtilis 168. Mol Microbiol 29(3):851–858
Stein T (2005) Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56(4):845–857
Steller S, Vater J (2000) Purification of the fengycin synthetase multienzyme system from Bacillus subtilis b213. J Chroma Togr B: Biomed Sci Appl 737(1):267–275
Steller S, Vollenbroich D, Leenders F, Stein T, Conrad B, Hofemeister J, Jacques P, Thonart P, Vater J (1999) Structural and functional organization of the fengycin synthetase multienzyme system from Bacillus subtilis b213 and A1/3. Chem Biol 6(1):31–41
Stragier P, Losick R (1996) Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30(1):297–341
Tabor CW, Tabor H (1984) Polyamines. Annu Rev Biochem 53(1):749–790
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acid Res 28(1):33–36
Tatusova TA, Madden TL (2006) BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174(2):247–250
Timmusk S, Grantcharova N, Wagner EGH (2005) Paenibacillus polymyxa invades plant roots and forms biofilms. Appl Environ Microbiol 71(11):7292
Tosato V, Albertini AM, Zotti M, Sonda S, Bruschi CV (1997) Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis 168. Microbiology 143(11):3443–3450
Walsh CT (2004) Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303(5665):1805
Weilharter A, Mitter B, Shin MV, Chain PS, Nowak J, Sessitsch A (2011) Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J Bacteriol 193(13):3383–3384
Werhane H, Lopez P, Mendel M, Zimmer M, Ordal G, Márquez-Magaña L (2004) The last gene of the fla/che operon in Bacillus subtilis, ylxL, is required for maximal σD function. J Bacteriol 186(12):4025–4029
Witte G, Hartung S, Buttner K, Hopfner KP (2008) Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell 30(2):167–178
Wolff S, Antelmann H, Albrecht D, Becher D, Bernhardt J, Bron S, Büttner K, van Dijl JM, Eymann C, Otto A (2007) Towards the entire proteome of the model bacterium Bacillus subtilis by gel-based and gel-free approaches. J Chroma Togr B 849(1–2):129–140
Wray L, Atkinson MR, Fisher SH (1994) The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII protein. J Bacteriol 176(1):108–114
**ong G, Zhao G, Fan C, He Y (2009) Identification and fungistatic effect of a biocontrol strain. J Yunnan Agr U 24:190–194
Acknowledgments
We thank the Ministry of Agriculture, China, and the Department of Science and Technologies, Yunnan Province, China, for their financial support through the Special Fund for Agroscientific Research in the Public Interest (201003029) and the Natural Science Foundation of Yunnan Province (2008CC024).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Standards
This article does not contain any studies with human participants or animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, S., Li, X., He, P. et al. Whole-genome sequencing of Bacillus subtilis XF-1 reveals mechanisms for biological control and multiple beneficial properties in plants. J Ind Microbiol Biotechnol 42, 925–937 (2015). https://doi.org/10.1007/s10295-015-1612-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1612-y