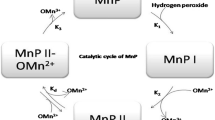
Abstract
DyP peroxidases comprise a novel superfamily of heme-containing peroxidases, which is unrelated to the superfamilies of plant and animal peroxidases. These enzymes have so far been identified in the genomes of fungi, bacteria, as well as archaea, although their physiological function is still unclear. DyPs are bifunctional enzymes displaying not only oxidative activity but also hydrolytic activity. Moreover, these enzymes are able to oxidize a variety of organic compounds of which some are poorly converted by established peroxidases, including dyes, β-carotene, and aromatic sulfides. Interestingly, accumulating evidence shows that microbial DyP peroxidases play a key role in the degradation of lignin. Owing to their unique properties, these enzymes are potentially interesting for a variety of biocatalytic applications. In this review, we deal with the biochemical and structural features of DyP-type peroxidases as well as their promising biotechnological potential.


Similar content being viewed by others
References
Adav SS, Ng CS, Arulmani M, Sze SK (2010) Quantitative iTRAQ secretome analysis of cellulolytic Thermobifida fusca. J Proteome Res 9:3016–3024
Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TD (2011) Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50:5096–5107
Amini S, Tavazoie S (2011) Antibiotics and the post-genome revolution. Curr Opin Microbiol 14:513–518
Banci L (1997) Structural properties of peroxidases. J Biotechnol 53:253–263
Brown ME, Barros T, Chang MC (2012) Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem Biol 7:2074–2081
Bugg TD, Ahmad M, Hardiman EM, Singh R (2011) The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol 22:394–400
Burner U, Krapfenbauer G, Furtmuller PG, Regelsberger G, Obinger C (2000) Oxidation of hydroquinone, 2,3-dimethylhydroquinone and 2,3,5-trimethylhydroquinone by human myeloperoxidase. Redox Rep 5:185–190
Chen H (2006) Recent advances in azo dye degrading enzyme research. Curr Protein Pept Sci 7:101–111
Choinowski T, Blodig W, Winterhalter KH, Piontek K (1999) The crystal structure of lignin peroxidase at 1.70-A resolution reveals a hydroxy group on the cbeta of tryptophan 171: a novel radical site formed during the redox cycle. J Mol Biol 286:809–827
Dailey HA et al (2011) The Escherichia coli protein YfeX functions as a porphyrinogen oxidase, not a heme dechelatase. MBio 2. doi:10.1128/mBio.00248-11
Davies MJ, Hawkins CL, Pattison DI, Rees MD (2008) Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxid Redox Signal 10:1199–1234
Doyle WA, Blodig W, Veitch NC, Piontek K, Smith AT (1998) Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Biochemistry 37:15097–15105
Faraco V, Piscitelli A, Sannia G, Giardina P (2007) Identification of a new member of the dye-decolorizing peroxidase family from Pleurotus ostreatus. World J Microbiol Biotechnol 23:889–893
Johjima T, Ohkuma M, Kudo T (2003) Isolation and cDNA cloning of novel hydrogen peroxide-dependent phenol oxidase from the basidiomycete Termitomyces albuminosus. Appl Microbiol Biotechnol 61:220–225
Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl JM (2004) Two minimal Tat translocases in Bacillus. Mol Microbiol 54:1319–1325
Kim SJ, Shoda M (1999) Purification and characterization of a novel peroxidase from Geotrichum candidum dec 1 involved in decolorization of dyes. Appl Environ Microbiol 65:1029–1035
Klibanov AM (2003) Asymmetric enzymatic oxidoreductions in organic solvents. Curr Opin Biotechnol 14:427–431
Kong L, Guo D, Zhou S, Yu X, Hou G, Li R, Zhao B (2010) Cloning and expression of a toxin gene from Pseudomonas fluorescens GcM5-1A. Arch Microbiol 192:585–593
Li J, Liu C, Li B, Yuan H, Yang J, Zheng B (2012) Identification and molecular characterization of a novel DyP-type peroxidase from Pseudomonas aeruginosa PKE117. Appl Biochem Biotechnol 166:774–785
Liers C, Bobeth C, Pecyna M, Ullrich R, Hofrichter M (2010) DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high-redox potential dyes. Appl Microbiol Biotechnol 85:1869–1879
Liers C, Pecyna MJ, Kellner H, Worrich A, Zorn H, Steffen KT, Hofrichter M, Ullrich R (2013) Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl Microbiol Biotechnol 97:5839–5849
Liers C, Ullrich R, Hofrichter M, Minibayeva FV, Beckett RP (2011) A heme peroxidase of the ascomyceteous lichen Leptogium saturninum oxidizes high-redox potential substrates. Fungal Genet Biol 48:1139–1145
Liu X et al (2011) Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes. J Biol Chem 286:14922–14931
Ogola HJ, Kamiike T, Hashimoto N, Ashida H, Ishikawa T, Shibata H, Sawa Y (2009) Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl Environ Microbiol 75:7509–7518
Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24:255–265
Puhse M, Szweda RT, Ma Y, Jeworrek C, Winter R, Zorn H (2009) Marasmius scorodonius extracellular dimeric peroxidase—exploring its temperature and pressure stability. Biochim Biophys Acta 1794:1091–1098
Regalado C, García-Almendárez BE, Duarte-Vázquez MA (2004) Biotechnological applications of peroxidases. Phytochem Rev 3:243–256
Reszka KJ, McCormick ML, Britigan BE (2001) Peroxidase- and nitrite-dependent metabolism of the anthracycline anticancer agents daunorubicin and doxorubicin. Biochemistry 40:15349–15361
Reszka KJ, Wagner BA, Burns CP, Britigan BE (2005) Effects of peroxidase substrates on the Amplex red/peroxidase assay: antioxidant properties of anthracyclines. Anal Biochem 342:327–337
Roberts JN, Singh R, Grigg JC, Murphy ME, Bugg TD, Eltis LD (2011) Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry 50:5108–5119
Salvachua D, Prieto A, Martinez AT, Martinez MJ (2013) Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl Environ Microbiol 79:4316–4324
Santos A, Mendes S, Brissos V, Martins LO (2013) New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl Microbiol Biotechnol. doi:10.1007/s00253-013-5041-4
Scheibner M, Hulsdau B, Zelena K, Nimtz M, de Boer L, Berger RG, Zorn H (2008) Novel peroxidases of Marasmius scorodonius degrade beta-carotene. Appl Microbiol Biotechnol 77:1241–1250
Sezer M, Santos A, Kielb P, Pinto T, Martins LO, Todorovic S (2013) Distinct structural and redox properties of the heme active site in bacterial dye decolorizing peroxidase-type peroxidases from two subfamilies: resonance Raman and electrochemical study. Biochemistry 52:3074–3084
Singh R, Grigg JC, Armstrong Z, Murphy ME, Eltis LD (2012) Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity. J Biol Chem 287:10623–10630
Stolz A (2001) Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 56:69–80
Strittmatter E, Liers C, Ullrich R, Wachter S, Hofrichter M, Plattner DA, Piontek K (2013) First crystal structure of a fungal high-redox potential dye-decolorizing peroxidase: substrate interaction sites and long-range electron transfer. J Biol Chem 288:4095–4102
Strittmatter E, Wachter S, Liers C, Ullrich R, Hofrichter M, Plattner DA, Piontek K (2013) Radical formation on a conserved tyrosine residue is crucial for DyP activity. Arch Biochem Biophys. doi:10.1016/j.abb.2013.07.007
Sturm A, Schierhorn A, Lindenstrauss U, Lilie H, Bruser T (2006) YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. J Biol Chem 281:13972–13978
Sugano Y (2009) DyP-type peroxidases comprise a novel heme peroxidase family. Cell Mol Life Sci 66:1387–1403
Sugano Y, Ishii Y, Shoda M (2004) Role of H164 in a unique dye-decolorizing heme peroxidase DyP. Biochem Biophys Res Commun 322:126–132
Sugano Y, Muramatsu R, Ichiyanagi A, Sato T, Shoda M (2007) DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J Biol Chem 282:36652–36658
Sutter M et al (2008) Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol 15:939–947
van Bloois E, Torres Pazmino DE, Winter RT, Fraaije MW (2010) A robust and extracellular heme-containing peroxidase from Thermobifida fusca as prototype of a bacterial peroxidase superfamily. Appl Microbiol Biotechnol 86:1419–1430
van Rantwijk F, Sheldon RA (2000) Selective oxygen transfer catalysed by heme peroxidases: synthetic and mechanistic aspects. Curr Opin Biotechnol 11:554–564
Veitch NC (2004) Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry 65:249–259
Welinder KG, Mauro JM, Norskov-Lauritsen L (1992) Structure of plant and fungal peroxidases. Biochem Soc Trans 20:337–340
Yoshida T, Tsuge H, Konno H, Hisabori T, Sugano Y (2011) The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue. FEBS J 278:2387–2394
Zamocky M, Jakopitsch C, Furtmuller PG, Dunand C, Obinger C (2008) The peroxidase-cyclooxygenase superfamily: reconstructed evolution of critical enzymes of the innate immune system. Proteins 72:589–605
Zubieta C et al (2007) Identification and structural characterization of heme binding in a novel dye-decolorizing peroxidase, TyrA. Proteins 69:234–243
Zubieta C et al (2007) Crystal structures of two novel dye-decolorizing peroxidases reveal a beta-barrel fold with a conserved heme-binding motif. Proteins 69:223–233
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Colpa, D.I., Fraaije, M.W. & van Bloois, E. DyP-type peroxidases: a promising and versatile class of enzymes. J Ind Microbiol Biotechnol 41, 1–7 (2014). https://doi.org/10.1007/s10295-013-1371-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1371-6