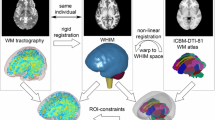
Abstract
Conventional brain injury metrics are scalars that treat the whole head/brain as a single unit but do not characterize the distribution of brain responses. Here, we establish a network-based “response feature matrix” to characterize the magnitude and distribution of impact-induced brain strains. The network nodes and edges encode injury risks to the gray matter regions and their white matter interconnections, respectively. The utility of the metric is illustrated in injury prediction using three independent, real-world datasets: two reconstructed impact datasets from the National Football League (NFL) and Virginia Tech, respectively, and measured concussive and non-injury impacts from Stanford University. Injury predictions with leave-one-out cross-validation are conducted using the two reconstructed datasets separately, and then by combining all datasets into one. Using support vector machine, the network-based injury predictor consistently outperforms four baseline scalar metrics including peak maximum principal strain of the whole brain (MPS), peak linear/rotational acceleration, and peak rotational velocity across all five selected performance measures (e.g., maximized accuracy of 0.887 vs. 0.774 and 0.849 for MPS and rotational acceleration with corresponding positive predictive values of 0.938, 0.772, and 0.800, respectively, using the reconstructed NFL dataset). With sufficient training data, real-world injury prediction is similar to leave-one-out in-sample evaluation, suggesting the potential advantage of the network-based injury metric over conventional scalar metrics. The network-based response feature matrix significantly extends scalar metrics by sampling the brain strains more completely, which may serve as a useful framework potentially allowing for other applications such as characterizing injury patterns or facilitating targeted multi-scale modeling in the future.









Similar content being viewed by others
References
Agoston DV, Langford D (2017) Big Data in traumatic brain injury; promise and challenges. Concussion (London, England) 2:CNC45. https://doi.org/10.2217/cnc-2016-0013
Bain AC, Meaney DF (2000) Tissue-level thresholds for axonal damage in an experimental model of central nervous system white matter injury. J Biomech Eng 122:615–622. https://doi.org/10.1115/1.1324667
Beckwith JG, Greenwald RM, Chu JJ (2012) Measuring head kinematics in football: correlation between the head impact telemetry system and Hybrid III headform. Ann Biomed Eng 40:237–248. https://doi.org/10.1007/s10439-011-0422-2
Beckwith JG, Greenwald RM, Chu JJ et al (2013) Head impact exposure sustained by football players on days of diagnosed Concussion. Med Sci Sports Exerc 45:737–746. https://doi.org/10.1249/MSS.0b013e3182792ed7
Beckwith JG, Zhao W, Ji S et al (2018) Estimated brain tissue response following impacts associated with and without diagnosed concussion. Ann Biomed Eng 46:819–830. https://doi.org/10.1007/s10439-018-1999-5
Bigler ED (2016) Systems biology, neuroimaging, neuropsychology, neuroconnectivity and traumatic brain injury. Front Syst Neurosci 10:1–23. https://doi.org/10.3389/FNSYS.2016.00055
Bigler ED, Maxwell WL (2012) Neuropathology of mild traumatic brain injury: relationship to neuroimaging findings. Brain Imaging Behav 6:108–136. https://doi.org/10.1007/s11682-011-9145-0
Cai Y, Wu S, Zhao W et al (2018) Concussion classification via deep learning using whole-brain white matter fiber strains. PLoS ONE 13:e0197992. https://doi.org/10.1371/journal.pone.0197992
Chen Y, Lin C (2006) Combining SVMs with various feature selection strategies. Strategies 324:1–10. https://doi.org/10.1007/978-3-540-35488-8_13
Cloots RJH (2011) Multi-scale mechanics of traumatic brain injury. Technische Universiteit Eindhoven
Cloots RJH, van Dommelen JAW, Kleiven S, Geers MGD (2012) Multi-scale mechanics of traumatic brain injury: predicting axonal strains from head loads. Biomech Model Mechanobiol 12:137–150. https://doi.org/10.1007/s10237-012-0387-6
Crisco JJ, Chu JJ, Greenwald RM (2004) An algorithm for estimating acceleration magnitude and impact location using multiple nonorthogonal single-axis accelerometers. J Biomech Eng 126:849–854
Duhaime A-C, Beckwith JG, Maerlender AC et al (2012) Spectrum of acute clinical characteristics of diagnosed concussions in college athletes wearing instrumented helmets. J Neurosurg 117:1092–1099. https://doi.org/10.3171/2012.8.JNS112298
Fornito A, Zalesky A, Bullmore E (eds) (2016) Fundamentals of brain network analysis. Elsevier/Academic Press Amsterdam, Boston
Ganpule S, Daphalapurkar NP, Ramesh KT et al (2017) A three-dimensional computational human head model that captures live human brain dynamics. J Neurotrauma 34:2154–2166. https://doi.org/10.1089/neu.2016.4744
Giordano C, Kleiven S (2014) Evaluation of axonal strain as a predictor for mild traumatic brain injuries using finite element modeling. Stapp Car Crash J 58:29–61
Giordano C, Kleiven S (2016) Development of an unbiased validation protocol to assess the biofidelity of finite element head models used in prediction of traumatic brain injury. Stapp Car Crash J 60:363–471
Giordano C, Zappalà S, Kleiven S (2017) Anisotropic finite element models for brain injury prediction: the sensitivity of axonal strain to white matter tract inter-subject variability. Biomech Model Mechanobiol 16:1269–1293. https://doi.org/10.1007/s10237-017-0887-5
Greenwald RM, Gwin JT, Chu JJ, Crisco JJ (2008) Head impact severity measures for evaluating mild traumatic brain injury risk exposure. Neurosurgery 62:789–798. https://doi.org/10.1227/01.neu.0000318162.67472.ad.Head
Hastie T, Tibshirani R, Friedman J (2008) The elements of statistical learning, data mining, inference, and prediction, 2nd edn. Springer, Berlin
Hernandez F, Wu LC, Yip MC et al (2015) Six degree-of-freedom measurements of human mild traumatic brain injury. Ann Biomed Eng 43:1918–1934. https://doi.org/10.1007/s10439-014-1212-4
Hernandez F, Giordano C, Goubran M et al (2019) Lateral impacts correlate with falx cerebri displacement and corpus callosum trauma in sports-related concussions. Biomech Model Mechanobiol. https://doi.org/10.1007/s10237-018-01106-0
Hira ZM, Gillies DF (2015) A review of feature selection and feature extraction methods applied on microarray data. Adv Bioinformatics 2015:198363. https://doi.org/10.1155/2015/198363
Ho J, Kleiven S (2009) Can sulci protect the brain from traumatic injury? J Biomech 42:2074–2080. https://doi.org/10.1016/j.jbiomech.2009.06.051
Ji S, Ghadyani H, Bolander R et al (2014) Parametric comparisons of intracranial mechanical responses from three validated finite element models of the human head. Ann Biomed Eng 42:11–24. https://doi.org/10.1007/s10439-013-0907-2
Ji S, Zhao W, Ford JC et al (2015) Group-wise evaluation and comparison of white matter fiber strain and maximum principal strain in sports-related concussion. J Neurotrauma 32:441–454. https://doi.org/10.1089/neu.2013.3268
Khvostikov A, Aderghal K, Benois-Pineau J, Krylov A, Catheline G (2018) 3D CNN-based classification using sMRI and MD-DTI images for Alzheimer disease studies. ar**v preprint ar**v:1801.05968
Kimpara H, Iwamoto M (2012) Mild traumatic brain injury predictors based on angular accelerations during impacts. Ann Biomed Eng 40:114–126. https://doi.org/10.1007/s10439-011-0414-2
King AIAI, Yang KHKH, Zhang L et al (2003) Is head injury caused by linear or angular acceleration? In: IRCOBI conference. Lisbon, Portugal, Portugal, pp 1–12
Kleiven S (2007) Predictors for traumatic brain injuries evaluated through accident reconstructions. Stapp Car Crash J 51:81–114
Koerte IK, Lin AP, Willems A et al (2015) A review of neuroimaging findings in repetitive brain trauma. Brain Pathol 25:318–349. https://doi.org/10.1111/bpa.12249
Kraft RH, Mckee PJ, Dagro AM, Grafton ST (2012) Combining the finite element method with structural connectome-based analysis for modeling neurotrauma: connectome neurotrauma mechanics. PLoS Comput Biol 8:e1002619. https://doi.org/10.1371/journal.pcbi.1002619
Kuo C, Wu L, Zhao W et al (2017) Propagation of errors from skull kinematic measurements to finite element tissue responses. Biomech Model Mechanobiol 17:235–247. https://doi.org/10.1007/s10237-017-0957-8
Leemans A, Jeurissen B, Siibers J, Jones D (2009) ExploreDTI: a graphical tool box for processing, analyzing, and visualizing diffusion MR data. In: 17th annual meeting of the international society of magnetic resonance in medicine. Hawaii, USA
Levin HS, Williams D, Crofford MJ et al (1988) Relationship of depth of brain lesions to consciousness and outcome after closed head injury. J Neurosurg 69:861–866. https://doi.org/10.3171/jns.1988.69.6.0861
Mao H, Zhang L, Jiang B et al (2013) Development of a finite element human head model partially validated with thirty five experimental cases. J Biomech Eng 135:111002–111015. https://doi.org/10.1115/1.4025101
Miller LE, Urban JE, Stitzel JD (2016) Development and validation of an atlas-based finite element brain model. Biomech Model 15:1201–1214. https://doi.org/10.1007/s10237-015-0754-1
Mohammadipour A, Alemi A (2017) Micromechanical analysis of brain’s diffuse axonal injury. J Biomech 65:61–74. https://doi.org/10.1016/j.jbiomech.2017.09.029
Montanino A, Kleiven S (2018) Utilizing a structural mechanics approach to assess the primary effects of injury loads onto the axon and its components. Front Neurol 9:643. https://doi.org/10.3389/fneur.2018.00643
Montenigro PH, Alosco ML, Martin BM et al (2017) Cumulative head impact exposure predicts later-life depression, apathy, executive dysfunction, and cognitive impairment in former high school and college football players. J Neurotrauma 13:55. https://doi.org/10.1089/neu.2016.4413
Newman JA, Beusenberg MC, Shewchenko N et al (2005) Verification of biomechanical methods employed in a comprehensive study of mild traumatic brain injury and the effectiveness of American football helmets. J Biomech 38:1469–1481. https://doi.org/10.1016/j.jbiomech.2004.06.025
NRC I (2014) Sports-related concussions in youth: improving the science, changing the culture. Washington, DC
Park HJ, Friston K (2013) Structural and functional brain networks: from connections to cognition. Science. https://doi.org/10.1126/science.1238411
Qi S, Meesters S, Nicolay K et al (2015) The influence of construction methodology on structural brain network measures: a review. J Neurosci Methods 253:170–182. https://doi.org/10.1016/j.jneumeth.2015.06.016
Rathore S, Habes M, Iftikhar MA et al (2017) A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. Neuroimage 155:530–548
Rowson B (2016) Evaluation and application of brain injury criteria to improve protective headgear design. Virginia Tech
Rowson S, Duma SM (2013) Brain injury prediction: assessing the combined probability of concussion using linear and rotational head acceleration. Ann Biomed Eng 41:873–882. https://doi.org/10.1007/s10439-012-0731-0
Rowson S, Duma SM, Beckwith JG et al (2012) Rotational head kinematics in football impacts: an injury risk function for concussion. Ann Biomed Eng 40:1–13. https://doi.org/10.1007/s10439-011-0392-4
Rowson S, Duma SM, Stemper BD et al (2018) Correlation of concussion symptom profile with head impact biomechanics: a case for individual-specific injury tolerance. J Neurotrauma 35:681–690. https://doi.org/10.1089/neu.2017.5169
Rowson S, Campolettano ET, Duma SM et al (2019) Accounting for variance in concussion tolerance between individuals: comparing head accelerations between concussed and physically matched control subjects. Ann Biomed Eng. https://doi.org/10.1007/s10439-019-02329-7
Sabet AA, Christoforou E, Zatlin B et al (2008) Deformation of the human brain induced by mild angular head acceleration. J Biomech 41:307–315. https://doi.org/10.1016/j.jbiomech.2007.09.016
Sahoo D, Deck C, Willinger R (2014) Development and validation of an advanced anisotropic visco-hyperelastic human brain FE model. J Mech Behav Biomed Mater 33:24–42. https://doi.org/10.1016/j.jmbbm.2013.08.022
Sahoo D, Deck C, Willinger R (2016) Brain injury tolerance limit based on computation of axonal strain. Accid Anal Prev 92:53–70. https://doi.org/10.1016/j.aap.2016.03.013
Sanchez EJ, Gabler LF, McGhee JS et al (2017) Evaluation of head and brain injury risk functions using sub-injurious human volunteer data. J Neurotrauma 34:2410–2424. https://doi.org/10.1089/neu.2016.4681
Sanchez EJ, Gabler LF, Good AB et al (2018) A reanalysis of football impact reconstructions for head kinematics and finite element modeling. Clin Biomech. https://doi.org/10.1016/j.clinbiomech.2018.02.019
Shattuck D, Mirza M, Adisetiyo V et al (2008) Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39:1064–1080
Šimundić A-M (2009) Measures of diagnostic accuracy: basic definitions. EJIFCC 19:203–211
Smith SM, Jenkinson M, Woolrich MW et al (2004) Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23:S208–S219
Sporns O (2013) Structure and function of complex brain networks. Dialogues Clin Neurosci 15:247–262
Stemper BD, Shah AS, Harezlak J et al (2018) Comparison of head impact exposure between concussed football athletes and matched controls: evidence for a possible second mechanism of sport-related concussion. Ann Biomed Eng. https://doi.org/10.1007/s10439-018-02136-6
Takhounts EG, Ridella SA, Tannous RE et al (2008) Investigation of traumatic brain injuries using the next generation of simulated injury monitor (SIMon) finite element head model. Stapp Car Crash J 52:1–31
Takhounts EGG, Craig MJJ, Moorhouse K et al (2013) Development of brain injury criteria (Br IC). Stapp Car Crash J 57:243–266
Versace J (1971) A review of the severity index. In: 15th stapp car crash conference. Coronado, CA, USA, p SAE paper 710881
Viano DC, Casson IR, Pellman EJ et al (2005) Concussion in professional football: brain responses by finite element analysis: part 9. Neurosurgery 57:891–915. https://doi.org/10.1227/01.NEU.0000186950.54075.3B
Wu LC, Nangia V, Bui K et al (2015) In vivo evaluation of wearable head impact sensors. Ann Biomed Eng 44:1234–1245. https://doi.org/10.1007/s10439-015-1423-3
Wu LC, Kuo C, Loza J et al (2018) Detection of american football head impacts using biomechanical features and support vector machine classification. Sci Rep 8:1–14. https://doi.org/10.1038/s41598-017-17864-3
Wu T, Alshareef A, Giudice JS, Panzer MB (2019) Explicit modeling of white matter axonal fiber tracts in a finite element brain model. Ann Biomed Eng. https://doi.org/10.1007/s10439-019-02239-8
Yanaoka T, Dokko Y, Takahashi Y (2015) Investigation on an injury criterion related to traumatic brain injury primarily induced by head rotation. SAE Tech Pap 2015-01-1439. https://doi.org/10.4271/2015-01-1439.copyright
Zhang L, Yang KHH, King AII (2004) A proposed injury threshold for mild traumatic brain injury. J Biomech Eng 126:226–236. https://doi.org/10.1115/1.1691446
Zhao W, Ji S (2017) Brain strain uncertainty due to shape variation in and simplification of head angular velocity profiles. Biomech Model Mechanobiol 16:449–461. https://doi.org/10.1007/s10237-016-0829-7
Zhao W, Ji S (2019a) Mesh convergence behavior and the effect of element integration of a human head injury model. Ann Biomed Eng 47:475–486. https://doi.org/10.1007/s10439-018-02159-z
Zhao W, Ji S (2019b) White matter anisotropy for impact simulation and response sampling in traumatic brain injury. J Neurotrauma 36:250–263. https://doi.org/10.1089/neu.2018.5634
Zhao W, Ford JC, Flashman LA et al (2016) White matter injury susceptibility via fiber strain evaluation using whole-brain tractography. J Neurotrauma 33:1834–1847. https://doi.org/10.1089/neu.2015.4239
Zhao W, Cai Y, Li Z, Ji S (2017) Injury prediction and vulnerability assessment using strain and susceptibility measures of the deep white matter. Biomech Model Mechanobiol 16:1709–1727. https://doi.org/10.1007/s10237-017-0915-5
Zhao W, Choate B, Ji S (2018) Material properties of the brain in injury-relevant conditions—experiments and computational modeling. J Mech Behav Biomed Mater 80:222–234. https://doi.org/10.1016/j.jmbbm.2018.02.005
Zhao W, Bartsch A, Benzel E et al (2019) Regional brain injury vulnerability in football from two finite element models of the human head. IRCOBI. Florence, Italy, pp 619–621
Zhou Z, Li X, Kleiven S, Hardy WN (2018) A reanalysis of experimental brain strain data: implication for finite element head model validation. Stapp Car Crash J 62:1–26
Zhu F, Gatti DL, Yang KH (2016) Nodal versus total axonal strain and the role of cholesterol in traumatic brain injury. J Neurotrauma 33:859–870. https://doi.org/10.1089/neu.2015.4007
Acknowledgements
Funding is provided by the NIH Grant R01 NS092853. The authors are grateful to Dr. David B. Camarillo at Stanford University for data sharing. They also thank Dr. Zheyang Wu at Worcester Polytechnic Institute for help on statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix: Out-of-sample prediction performance
Appendix: Out-of-sample prediction performance
The network-based injury metric was retrained using the entire NFL or VT dataset. Tables 4 and 5 in “Appendix” report the out-of-sample performances when predicting injury occurrences for impacts in the other reconstructed and SF dataset. For each injury predictor trained on the same training dataset, the injury prediction performance depended on the testing dataset used. For example, the network-based metric trained on the NFL dataset to maximize accuracy achieved a rather poor sensitivity with VT dataset (0.091, i.e., only one of the 11 concussion cases was correctly predicted), but a perfect score of 1.000 for the SF dataset (both concussions correctly predicted).
On the other hand, when using the same SF dataset for testing, the performance of each injury metric in most categories also depended on the training dataset used. For example, the best performer in terms of out-of-sample prediction accuracy was \(v_{\text{rot}}\) when trained on the NFL dataset, but it was \(a_{\text{lin}}\) when trained on the VT dataset (accuracy of 0.936 vs. 0.927; Tables 4 and 5 in “Appendix”). The dependency on the training dataset was also evident when comparing the injury thresholds established. Using those from the NFL dataset as baselines, the injury thresholds for MPS, \(a_{\text{lin}}\), \(a_{\text{rot}}\), and \(v_{\text{rot}}\) differed by − 24.2%, 28.8%, − 9.9%, − 18.3%, respectively.
Interestingly, the majority of “best performers” were baseline kinematic variables across many categories. For example, \(a_{\text{rot}}\) trained on the NFL dataset achieved the best prediction accuracy and testing AUC for the VT dataset (Table 4), and the same variable trained on VT dataset also achieved the best accuracy and AUC when predicting the NFL impacts (Table 5). However, the same predictor did not receive the best accuracy or AUC when predicting impacts in the SF dataset, regardless of which training dataset used. This observation confirmed that the injury prediction performance depended on the testing dataset. Further, kinematic variables could also yield the worst performances. For example, training \(a_{\text{lin}}\) based on the VT dataset led to a sensitivity of zero when predicting injury in the SF dataset (i.e., none of the two concussions were correctly predicted).
To summarize, these findings suggest that the performance of a specific injury predictor depends not only on the injury metric itself, but also on the training and testing datasets as well. Therefore, out-of-sample injury prediction is effectively a “three-way” fitting process.
Rights and permissions
About this article
Cite this article
Wu, S., Zhao, W., Rowson, B. et al. A network-based response feature matrix as a brain injury metric. Biomech Model Mechanobiol 19, 927–942 (2020). https://doi.org/10.1007/s10237-019-01261-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10237-019-01261-y