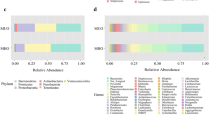
Abstract
Aims
To identify fecal microbiota profiles associated with metabolic abnormalities belonging to the metabolic syndrome (MS), high count of white blood cells (WBCs) and insulin resistance (IR).
Methods
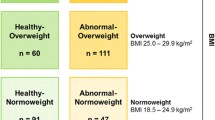
Sixty-eight young patients with obesity were stratified for percentile distribution of MS abnormalities. A MS risk score was defined as low, medium, and high MS risk. High WBCs were defined as a count ≥ 7.0 103/µL; severe obesity as body mass index Z-score ≥ 2 standard deviations; IR as homeostatic assessment model algorithm of IR (HOMA) ≥ 3.7. Stool samples were analyzed by 16S rRNA-based metagenomics.
Results
We found reduced bacterial richness of fecal microbiota in patients with IR and high diastolic blood pressure (BP). Distinct microbial markers were associated to high BP (Clostridium and Clostridiaceae), low high-density lipoprotein cholesterol (Lachnospiraceae, Gemellaceae, Turicibacter), and high MS risk (Coriobacteriaceae), WBCs (Bacteroides caccae, Gemellaceae), severe obesity (Lachnospiraceae), and impaired glucose tolerance (Bacteroides ovatus and Enterobacteriaceae). Conversely, taxa such as Faecalibacterium prausnitzii, Parabacterodes, Bacteroides caccae, Oscillospira, Parabacterodes distasonis, Coprococcus, and Haemophilus parainfluenzae were associated to low MS risk score, triglycerides, fasting glucose and HOMA-IR, respectively. Supervised multilevel analysis grouped clearly “variable” patients based on the MS risk.
Conclusions
This was a proof-of-concept study opening the way at the identification of fecal microbiota signatures, precisely associated with cardiometabolic risk factors in young patients with obesity. These evidences led us to infer, while some gut bacteria have a detrimental role in exacerbating metabolic risk factors some others are beneficial ameliorating cardiovascular host health.






Similar content being viewed by others
Availability of data and materials
Sequencing reads and the associated metadata are available at BioProject database of NCBI (PRJNA356507 and PRJNA280490) (https://www.ncbi.nlm.nih.gov/bioproject/).
Abbreviations
- 2HPG:
-
2H plasma glucose
- ALT:
-
Alanine aminotransferase
- AST:
-
Aspartate aminotransferase
- BMI:
-
Body mass index
- CVD:
-
Cardiovascular disease
- DBP:
-
Diastolic blood pressures
- HbA1c:
-
Hemoglobin A1c
- HDL-C:
-
High-density lipoprotein cholesterol
- HFD:
-
High-fat diet
- HOMA-IR:
-
Homeostatic assessment model algorithm of insulin resistance
- IGT:
-
Impaired glucose tolerance
- IQR:
-
Interquartile range
- IR:
-
Insulin resistance
- LDA:
-
Linear discriminant analysis
- LEfSe:
-
Linear discriminant analysis effect size
- MS:
-
Metabolic syndrome
- NAST:
-
Nearest Alignment Space Termination
- NGT:
-
Normal glucose tolerance
- OPBG:
-
''Bambino Gesù'' Children’s Hospital
- OTUs:
-
Operational taxonomic units
- PCA:
-
Principal component analysis
- PCoA:
-
Principal coordinate analysis
- PLS:
-
Sparse partial least squares
- QIIME:
-
Quantitative Insights Into Microbial Ecology
- SBP:
-
Systolic blood pressures
- SDS:
-
Standard deviation score
- T2D:
-
Type 2 diabetes
- WBCs:
-
White blood cells
- γ-GT:
-
γ-Glutamyl transferase
References
Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C et al (2014) Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 384:766–781
Manco M (2011) Metabolic syndrome in childhood from impaired carbohydrate metabolism to nonalcoholic fatty liver disease. J Am Coll Nutr 30:295–303
Alshehri AM (2010) Metabolic syndrome and cardiovascular risk. J Family Community Med 17:73–78
Organization WH (2016) Consideration of the evidence on childhood obesity for the Commission on Ending Childhood Obesity: report of the ad hoc working group on science and evidence for ending childhood obesity. Switzerland, Geneva
Manco M, Putignani L, Bottazzo GF (2010) Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev 31:817–844
Tremaroli V, Bäckhed F (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249
Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H (2018) Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr Rev 39:133–153
Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR (2008) Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci 105:13580–13585
Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B et al (2011) Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63
Wolf AJ, Underhill DM (2018) Peptidoglycan recognition by the innate immune system. Nat Rev Immunol 18:243–254
Cole TJ, Bellizzi MC, Flegal KM, Dietz WH (2000) Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ 320:1240–1243
Podoll A, Grenier M, Croix B, Feig DI (2007) Inaccuracy in pediatric outpatient blood pressure measurement. Pediatrics 119:e538–e543
Shashaj B, Luciano R, Contoli B, Morino GS, Spreghini MR, Rustico C et al (2016) Reference ranges of HOMA-IR in normal-weight and obese young Caucasians. Acta Diabetol 53:251–260
Tanner JM, Whitehouse RH (1976) Clinical longitudinal standards for height, weight, height velocity, weight velocity, and stages of puberty. Arch Dis Child 51:170–179
Martino F, Puddu PE, Pannarale G, Colantoni C, Zanoni C, Martino E et al (2014) Metabolic syndrome among children and adolescents from Southern Italy: contribution from the Calabrian Sierras Community Study (CSCS). Int J Cardiol 177:455–460
Spreghini N, Cianfarani S, Spreghini MR, Brufani C, Morino GS, Inzaghi E et al (2019) Oral glucose effectiveness and metabolic risk in obese children and adolescents. Acta Diabetol 56:955–962
Wallace TM, Levy JC, Matthews DR (2004) Use and abuse of HOMA modeling. Diabetes Care 27:1487–1495
Nilsson G, Hedberg P, Öhrvik J (2014) White blood cell count in elderly is clinically useful in predicting long-term survival. J Aging Res 2014:1–6
Del Chierico F, Nobili V, Vernocchi P, Russo A, De Stefanis C, Gnani D et al (2017) Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65:451–464
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al (2006) NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34:W394-399
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS et al (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Maitra S, Yan J (2008) Principle component analysis and partial least squares: two dimension reduction techniques for regression. Appl Multivar Stat Models 79:79–90
Del Chierico F, Abbatini F, Russo A, Quagliariello A, Reddel S, Capoccia D, et al. Gut Microbiota Markers in Obese Adolescent and Adult Patients: Age-Dependent Differential Patterns. Frontiers in Microbiology [Internet]. 2018 [cited 2020 Jan 23];9. Available from: https://www.frontiersin.org/article/https://doi.org/10.3389/fmicb.2018.01210/full
Yuan X, Chen R, Zhang Y, Lin X, Yang X. Gut Microbiota: Effect of Pubertal Status [Internet]. In Review; 2020 Aug. Available from: https://www.researchsquare.com/article/rs-52945/v1
Yuan X, Chen R, Zhang Y, Lin X, Yang X. Sexual dimorphism of gut microbiota at different pubertal status. Microbial Cell Factories [Internet]. 2020 [cited 2020 Oct 9];19. Available from: https://microbialcellfactories.biomedcentral.com/articles/https://doi.org/10.1186/s12934-020-01412-2
Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G et al (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500:541
Underwood MA (2014) Intestinal dysbiosis: Novel mechanisms by which gut microbes trigger and prevent disease. Prev Med 65:133–137
Sun S, Lulla A, Sioda M, Winglee K, Wu MC, Jacobs DR et al (2019) Gut microbiota composition and blood pressure:the CARDIA Study. Hypertension 73:998–1006
Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM et al (2015) Gut dysbiosis is linked to hypertension. Hypertension 65:1331–1340
Bhute SS, Ghaskadbi SS, Shouche YS. Rare Biosphere in Human Gut: A Less Explored Component of Human Gut Microbiota and Its Association with Human Health. In: Kalia VC, Shouche Y, Purohit HJ, Rahi P, editors. Mining of Microbial Wealth and MetaGenomics [Internet]. Singapore: Springer Singapore; 2017 [cited 2020 Jun 12]. p. 133–42. Available from: http://springer.longhoe.net/https://doi.org/10.1007/978-981-10-5708-3_8
Granado-Serrano AB, Martín-Garí M, Sánchez V, Solans MR, Berdún R, Ludwig IA et al (2019) Faecal bacterial and short-chain fatty acids signature in hypercholesterolemia. Sci Rep 9:1–13
Braun T, Di Segni A, BenShoshan M, Neuman S, Levhar N, Bubis M et al (2019) Individualized dynamics in the gut microbiota precede Crohnʼs disease flares. Am J Gastroenterol 114:1142–1151
Zeze K, Hirano A, Torisu T, Esaki M, Shibata H, Moriyama T et al (2020) Mucosal dysbiosis in patients with gastrointestinal follicular lymphoma. Hematol Oncol 38:181–188
Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH et al (2014) Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J 8:2218–2230
Wang K, Liao M, Zhou N, Bao L, Ma K, Zheng Z et al (2019) Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep 26(222–235):e5
Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano G, Gasbarrini A et al (2019) What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 7:14
Wexler HM (2007) Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20:593–621
Chen Y, Zheng H, Zhang G, Chen F, Chen L, Yang Z. High oscillospira abundance indicates constipation and low BMI in the Guangdong Gut Microbiome Project. Sci Rep [Internet]. 2020 [cited 2020 Jul 16];10. Available from: http://www.nature.com/articles/s41598-020-66369-z
Liu H, Zhang H, Wang X, Yu X, Hu C, Zhang X (2018) The family Coriobacteriaceae is a potential contributor to the beneficial effects of Roux-en-Y gastric bypass on type 2 diabetes. Surgery for Obesity and Related Diseases 14:584–593
Gao Y, Yang L, Chin Y, Liu F, Li RW, Yuan S et al (2020) Astaxanthin n-octanoic acid diester ameliorates insulin resistance and modulates gut microbiota in high-fat and high-sucrose diet-fed mice. Int J Mol Sci 21:2149
Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. Federici M, editor. PLoS ONE. 2013;8:e71108
Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. Bereswill S, editor. PLoS ONE. 2010;5:e9085
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F et al (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S et al (2010) Impact of diet in sha** gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci 107:14691–14696
Munukka E, Rintala A, Toivonen R, Nylund M, Yang B, Takanen A et al (2017) Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J Nat 11:1667–1679
Brambilla P, Lissau I, Flodmark C-E, Moreno LA, Widhalm K, Wabitsch M et al (2007) Metabolic risk-factor clustering estimation in children: to draw a line across pediatric metabolic syndrome. Int J Obes (Lond) 31:591–600
Acknowledgments
None.
Funding
This work was supported by the European Commission under the 7th FP_Information Communication Technologies Programme_MD-Paedigree, Model Driven Paediatric European Digital Repository, (Grant Agreement No. 600932).
Author information
Authors and Affiliations
Contributions
Conceptualization, Melania Manco; Data curation, Marzia Bianchi; Formal analysis, Federica Del Chierico, Valentina Tortosa and Andrea Quagliariello, Simone Gardini, Valerio Guarrasi; Funding acquisition, Melania Manco; Investigation, Alessandra Russo, Blegina Shashaj and Danilo Fintini; Methodology, Alessandra Russo; Project administration, Marzia Bianchi; Resources, Danilo Fintini and Lorenza Putignani; Software, Valentina Tortosa, Andrea Quagliariello, Simone Gardini, Valerio Guarrasi; Supervision, Lorenza Putignani; Writing – original draft, Federica Del Chierico and Melania Manco; Writing – review & editing, Federica Del Chierico, Melania Manco, Alessandra Russo, Marzia Bianchi, Valentina Tortosa, Andrea Quagliariello, Simone Gardini, Valerio Guarrasi, Blegina Shashaj, Danilo Fintini and Lorenza Putignani. All authors had final approval of the submitted and published versions.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Ethics approval
The study was approved by the OPBG ethical committee (protocol #615/2013) and was conducted in accordance with the Principles of Good Clinical Practice and the Declaration of Helsinki.
Informed consent
Written informed consent was obtained from all participants.
Additional information
This article belongs to the topical collection Gut Microbiome and Metabolic Disorders, managed by Massimo Federici.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Del Chierico, F., Manco, M., Gardini, S. et al. Fecal microbiota signatures of insulin resistance, inflammation, and metabolic syndrome in youth with obesity: a pilot study. Acta Diabetol 58, 1009–1022 (2021). https://doi.org/10.1007/s00592-020-01669-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00592-020-01669-4